IsomaltoheptaoseCAS# 6513-12-8 |
Quality Control & MSDS
Number of papers citing our products

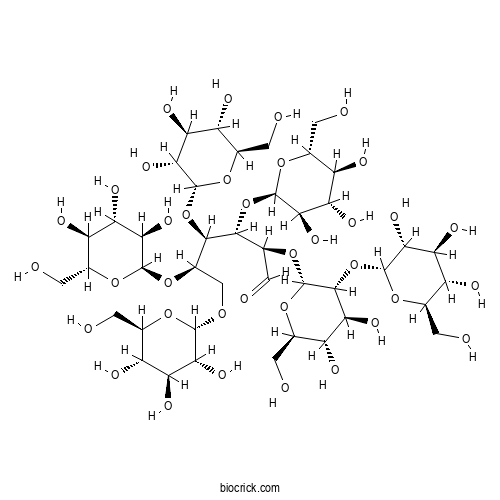
Chemical structure

3D structure
| Cas No. | 6513-12-8 | SDF | Download SDF |
| PubChem ID | 3081403.0 | Appearance | Powder |
| Formula | C42H72O36 | M.Wt | 1153.0 |
| Type of Compound | Oligoses | Storage | Desiccate at -20°C |
| Solubility | Soluble in Chloroform,Dichloromethane,Ethyl Acetate,DMSO,Acetone,etc. | ||
| Chemical Name | (2R,3S,4R,5R)-2-[(2R,3R,4S,5S,6R)-4,5-dihydroxy-6-(hydroxymethyl)-3-[(2R,3R,4S,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxyoxan-2-yl]oxy-3,4,5,6-tetrakis[[(2R,3R,4S,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy]hexanal | ||
| SMILES | C(C1C(C(C(C(O1)OCC(C(C(C(C=O)OC2C(C(C(C(O2)CO)O)O)OC3C(C(C(C(O3)CO)O)O)O)OC4C(C(C(C(O4)CO)O)O)O)OC5C(C(C(C(O5)CO)O)O)O)OC6C(C(C(C(O6)CO)O)O)O)O)O)O)O | ||
| Standard InChIKey | YGMBQDCBGPAZNW-YIBJATESSA-N | ||
| Standard InChI | InChI=1S/C42H72O36/c43-1-9-17(50)23(56)29(62)37(68-9)67-8-16(75-38-30(63)24(57)18(51)10(2-44)69-38)35(77-40-32(65)26(59)20(53)12(4-46)71-40)34(76-39-31(64)25(58)19(52)11(3-45)70-39)15(7-49)74-42-36(28(61)22(55)14(6-48)73-42)78-41-33(66)27(60)21(54)13(5-47)72-41/h7,9-48,50-66H,1-6,8H2/t9-,10-,11-,12-,13-,14-,15+,16-,17-,18-,19-,20-,21-,22-,23+,24+,25+,26+,27+,28+,29-,30-,31-,32-,33-,34-,35-,36-,37+,38-,39-,40-,41-,42-/m1/s1 | ||
| General tips | For obtaining a higher solubility , please warm the tube at 37 ℃ and shake it in the ultrasonic bath for a while.Stock solution can be stored below -20℃ for several months. We recommend that you prepare and use the solution on the same day. However, if the test schedule requires, the stock solutions can be prepared in advance, and the stock solution must be sealed and stored below -20℃. In general, the stock solution can be kept for several months. Before use, we recommend that you leave the vial at room temperature for at least an hour before opening it. |
||
| About Packaging | 1. The packaging of the product may be reversed during transportation, cause the high purity compounds to adhere to the neck or cap of the vial.Take the vail out of its packaging and shake gently until the compounds fall to the bottom of the vial. 2. For liquid products, please centrifuge at 500xg to gather the liquid to the bottom of the vial. 3. Try to avoid loss or contamination during the experiment. |
||
| Shipping Condition | Packaging according to customer requirements(5mg, 10mg, 20mg and more). Ship via FedEx, DHL, UPS, EMS or other couriers with RT, or blue ice upon request. | ||

Isomaltoheptaose Dilution Calculator

Isomaltoheptaose Molarity Calculator
| 1 mg | 5 mg | 10 mg | 20 mg | 25 mg | |
| 1 mM | 0.8673 mL | 4.3365 mL | 8.673 mL | 17.3461 mL | 21.6826 mL |
| 5 mM | 0.1735 mL | 0.8673 mL | 1.7346 mL | 3.4692 mL | 4.3365 mL |
| 10 mM | 0.0867 mL | 0.4337 mL | 0.8673 mL | 1.7346 mL | 2.1683 mL |
| 50 mM | 0.0173 mL | 0.0867 mL | 0.1735 mL | 0.3469 mL | 0.4337 mL |
| 100 mM | 0.0087 mL | 0.0434 mL | 0.0867 mL | 0.1735 mL | 0.2168 mL |
| * Note: If you are in the process of experiment, it's necessary to make the dilution ratios of the samples. The dilution data above is only for reference. Normally, it's can get a better solubility within lower of Concentrations. | |||||

Calcutta University

University of Minnesota

University of Maryland School of Medicine

University of Illinois at Chicago

The Ohio State University

University of Zurich

Harvard University

Colorado State University

Auburn University

Yale University

Worcester Polytechnic Institute

Washington State University

Stanford University

University of Leipzig

Universidade da Beira Interior

The Institute of Cancer Research

Heidelberg University

University of Amsterdam

University of Auckland

TsingHua University

The University of Michigan

Miami University

DRURY University

Jilin University

Fudan University

Wuhan University

Sun Yat-sen University

Universite de Paris

Deemed University

Auckland University

The University of Tokyo

Korea University
- Isomaltooctaose
Catalog No.:BCX1008
CAS No.:35867-37-9
- Alitame hydrate
Catalog No.:BCX1007
CAS No.:99016-42-9
- Euphorbetin
Catalog No.:BCX1006
CAS No.:35897-99-5
- Hericene A
Catalog No.:BCX1005
CAS No.:157207-54-0
- Hericene B
Catalog No.:BCX1004
CAS No.:157207-55-1
- α-Glucosyl Hesperidin
Catalog No.:BCX1003
CAS No.:161713-86-6
- Neohesperidose
Catalog No.:BCX1002
CAS No.:17074-02-1
- Ovatodiolide
Catalog No.:BCX1001
CAS No.:3484-37-5
- Anisomelic acid
Catalog No.:BCX1000
CAS No.:59632-76-7
- Apigenin triacetate
Catalog No.:BCX0999
CAS No.:3316-46-9
- Homodihydrocapsaicin II
Catalog No.:BCX0998
CAS No.:71239-21-9
- Neopuerarin B
Catalog No.:BCX0997
CAS No.:1150314-39-8
- Isomaltohexaose
Catalog No.:BCX1010
CAS No.:6175-02-6
- Isomaltopentaose
Catalog No.:BCX1011
CAS No.:6082-32-2
- Isomaltotetraose
Catalog No.:BCX1012
CAS No.:35997-20-7
- 6'-Hydroxy-3,4,2',3',4'-pentamethoxychalcone
Catalog No.:BCX1013
CAS No.:114021-62-4
- Cycloastragenol-6-O-β-D-glucoside
Catalog No.:BCX1014
CAS No.:86764-12-7
- 6,8-Dihydroxy-1,2,7-trimethoxy-3-methylanthraquinone
Catalog No.:BCX1015
CAS No.:1622982-59-5
- Zeatin
Catalog No.:BCX1016
CAS No.:13114-27-7
- Neopuerarin A
Catalog No.:BCX1017
CAS No.:1150314-34-3
- Docosyl ferulate
Catalog No.:BCX1018
CAS No.:101927-24-6
- Furopelargone B
Catalog No.:BCX1019
CAS No.:1143-46-0
- Pseudostellarin A
Catalog No.:BCX1020
CAS No.:156430-20-5
- Soyasaponin Be
Catalog No.:BCX1021
CAS No.:117210-14-7
A novel intracellular dextranase derived from Paenibacillus sp. 598K with an ability to degrade cycloisomaltooligosaccharides.[Pubmed:31273396]
Appl Microbiol Biotechnol. 2019 Aug;103(16):6581-6592.
Paenibacillus sp. 598K produces cycloisomaltooligosaccharides (CIs) in culture from dextran and starch. CIs are cyclic oligosaccharides consisting of seven or more alpha-(1 --> 6)-linked-D-glucose residues. The extracellular enzyme CI glucanotransferase (PsCITase), which is the member of glycoside hydrolase family 66, catalyzes the final stage of CI production and produces mainly cycloIsomaltoheptaose. We have discovered a novel intracellular CI-degrading dextranase (PsDEX598) from Paenibacillus sp. 598K. The 69.7-kDa recombinant PsDEX598 does not digest isomaltotetraose or shorter isomaltooligosaccharides, but digests longer ones of at least up to Isomaltoheptaose. It also digests oligoCIs of cycloIsomaltoheptaose, cycloisomaltooctaose, and cycloisomaltononaose better than it does with megaloCIs of cycloisomaltodecaose, cycloisomaltoundecaose, and cycloisomaltododecaose, as well as an alpha-(1 --> 6)-D-glucan of dextran 40. PsDEX598 is produced intracellularly when culture medium is supplemented with cycloIsomaltoheptaose or dextran, but not with isomaltooligosaccharides (a mixture of isomaltose, isomaltotriose, and panose), starch, or glucose. The whole genomic DNA sequence of the strain 598K implies that it harbors two genes for enzymes belonging to glycoside hydrolase family 66 (PsCITase and PsDEX598), and PsDEX598 is the only dextranase in the strain. PsDEX598 does not have any carbohydrate-binding modules (CBMs) and has a low similarity (< 30%) with other family 66 dextranases, and the catalytic amino acids of this enzyme are predicted to be Asp191, Asp303, and Glu368. The strain Paenibacillus sp. 598K appears to take up CI-7, so these findings indicate that this bacterium can degrade CIs using a dextranase within the cells and so utilize them as a carbon source for growth.
Isomaltooligosaccharide-binding structure of Paenibacillus sp. 598K cycloisomaltooligosaccharide glucanotransferase.[Pubmed:28385816]
Biosci Rep. 2017 Apr 28;37(2):BSR20170253.
Paenibacillus sp. 598K cycloisomaltooligosaccharide glucanotransferase (CITase), a member of glycoside hydrolase family 66 (GH66), catalyses the intramolecular transglucosylation of dextran to produce CIs with seven or more degrees of polymerization. To clarify the cyclization reaction and product specificity of the enzyme, we determined the crystal structure of PsCITase. The core structure of PsCITase consists of four structural domains: a catalytic (beta/alpha)(8)-domain and three beta-domains. A family 35 carbohydrate-binding module (first CBM35 region of Paenibacillus sp. 598K CITase, (PsCBM35-1)) is inserted into and protrudes from the catalytic domain. The ligand complex structure of PsCITase prepared by soaking the crystal with cycloIsomaltoheptaose yielded bound sugars at three sites: in the catalytic cleft, at the joint of the PsCBM35-1 domain and at the loop region of PsCBM35-1. In the catalytic site, soaked cycloIsomaltoheptaose was observed as a linear Isomaltoheptaose, presumably a hydrolysed product from cycloIsomaltoheptaose by the enzyme and occupied subsites -7 to -1. Beyond subsite -7, three glucose moieties of another isomaltooiligosaccharide were observed, and these positions are considered to be distal subsites -13 to -11. The third binding site is the canonical sugar-binding site at the loop region of PsCBM35-1, where the soaked cycloIsomaltoheptaose is bound. The structure indicated that the concave surface between the catalytic domain and PsCBM35-1 plays a guiding route for the long-chained substrate at the cyclization reaction.
Structural elucidation of the cyclization mechanism of alpha-1,6-glucan by Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase.[Pubmed:24616103]
J Biol Chem. 2014 Apr 25;289(17):12040-12051.
Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase belongs to the glycoside hydrolase family 66 and catalyzes an intramolecular transglucosylation reaction that produces cycloisomaltooligosaccharides from dextran. The crystal structure of the core fragment from Ser-39 to Met-738 of B. circulans T-3040 cycloisomaltooligosaccharide glucanotransferase, devoid of its N-terminal signal peptide and C-terminal nonconserved regions, was determined. The structural model contained one catalytic (beta/alpha)8-barrel domain and three beta-domains. Domain N with an immunoglobulin-like beta-sandwich fold was attached to the N terminus; domain C with a Greek key beta-sandwich fold was located at the C terminus, and a carbohydrate-binding module family 35 (CBM35) beta-jellyroll domain B was inserted between the 7th beta-strand and the 7th alpha-helix of the catalytic domain A. The structures of the inactive catalytic nucleophile mutant enzyme complexed with isomaltohexaose, Isomaltoheptaose, isomaltooctaose, and cycloisomaltooctaose revealed that the ligands bound in the catalytic cleft and the sugar-binding site of CBM35. Of these, isomaltooctaose bound in the catalytic site extended to the second sugar-binding site of CBM35, which acted as subsite -8, representing the enzyme.substrate complex when the enzyme produces cycloisomaltooctaose. The Isomaltoheptaose and cycloisomaltooctaose bound in the catalytic cleft with a circular structure around Met-310, representing the enzyme.product complex. These structures collectively indicated that CBM35 functions in determining the size of the product, causing the predominant production of cycloisomaltooctaose by the enzyme. The canonical sugar-binding site of CBM35 bound the mid-part of isomaltooligosaccharides, indicating that the original function involved substrate binding required for efficient catalysis.
Biochemical characterization of a novel cycloisomaltooligosaccharide glucanotransferase from Paenibacillus sp. 598K.[Pubmed:22542750]
Biochim Biophys Acta. 2012 Jul;1824(7):919-24.
Cycloisomaltooligosaccharide glucanotransferase (CITase; EC 2.4.1.248), a member of the glycoside hydrolase family 66 (GH66), catalyzes the intramolecular transglucosylation of dextran to produce cycloisomaltooligosaccharides (CIs; cyclodextrans) of varying lengths. Eight CI-producing bacteria have been found; however, CITase from Bacillus circulans T-3040 (CITase-T3040) is the only CI-producing enzyme that has been characterized to date. In this study, we report the gene cloning, enzyme characterization, and analysis of essential Asp and Glu residues of a novel CITase from Paenibacillus sp. 598K (CITase-598K). The cit genes from T-3040 and 598K strains were expressed recombinantly, and the properties of Escherichia coli recombinant enzymes were compared. The two CITases exhibited high primary amino acid sequence identity (67%). The major product of CITase-598K was cycloIsomaltoheptaose (CI-7), whereas that of CITase-T3040 was cycloisomaltooctaose (CI-8). Some of the properties of CITase-598K are more favorable for practical use compared with CITase-T3040, i.e., the thermal stability for CITase-598K (Isomaltoheptaose or smaller substrates were used, a lag time was observed before the intramolecular transglucosylation reaction began. As substrate length increased, the lag time shortened. Catalytically important residues of CITase-598K were predicted to be Asp144, Asp269, and Glu341. These findings will serve as a basis for understanding the reaction mechanism and substrate recognition of GH66 enzymes.
Single-molecule force spectroscopy for studying kinetics of enzymatic dextran elongations.[Pubmed:21443256]
J Am Chem Soc. 2011 Apr 20;133(15):5701-3.
Catalytic elongation of dextran by a single molecule of dextransucrase (DSase) was directly monitored by observing the movements of the positions of a rupture peak, which represented the adhesive force between an Isomaltoheptaose (dextran 7-mer)-immobilized probe and a DSase-immobilized mica surface. This was initiated with the addition of sucrose monomers. From the histograms of the rupture peaks after elongation reactions on each individual enzyme and the continuous peak shift of certain single enzymes, the catalytic elongation rate constant (k(cat)) was ascertained to be 1.2-2.7 s(-1).
Comparison of the glass transition temperature and fragility parameter of isomalto-olygomer predicted by molecular dynamics simulations with those measured by differential scanning calorimetry.[Pubmed:16272728]
Chem Pharm Bull (Tokyo). 2005 Nov;53(11):1443-5.
The purpose of this study is to examine whether molecular dynamics (MD) simulations using a commercially available software for personal computers can estimate the glass transition temperature (Tg) of amorphous systems containing pharmaceutically-relevant excipients. MD simulations were carried out with an amorphous matrix model constructed from Isomaltoheptaose, and the Tg estimated from the calculated density versus temperature profile was compared with the Tg measured by differential scanning calorimetry (DSC) for freeze-dried isomalto-oligomer having an average molecular weight close to that of Isomaltoheptaose. The Tg values determined by DSC were lower by 10 to 20 K than those extrapolated from the Tg values estimated by MD simulation. Fragility parameter was estimated to be 56 and 51 from MD simulation and from DSC measurement, respectively. Thus, the results suggest that MD simulation can provide approximate estimates for the Tg and fragility parameter of amorphous formulations. However, a reduction of the cooling rate, achievable by sufficiently elongating the simulation duration, is necessary for more accurate estimation.
Altering the substrate chain-length specificity of an alpha-glucosidase.[Pubmed:12727208]
Biochem Biophys Res Commun. 2003 May 16;304(4):684-90.
Dextran glucosidases show high sequence identity (50%) to Bacillus sp. SAM1606 alpha-glucosidase, which is more specific for short-chain substrates. Sequence comparison of these enzymes as well as molecular modeling studies predicted that the extension of loop 4 of the (beta/alpha)(8)-barrel fold may be responsible for the narrower specificity of SAM1606 alpha-glucosidase with respect to substrate chain length. Indeed, deletion mutants of SAM1606 alpha-glucosidase that lack this extension showed higher relative activities toward dextran and long-chain isomaltooligosaccharides. Kinetic and thermodynamic analyses of oligosaccharide hydrolysis catalyzed by SAM1606 alpha-glucosidase and its deletion mutants suggested that the loss of such extension(s) in loop 4 should energetically destabilize the Michaelis complexes with long-chain substrates to result in smaller differences between the activation free energies for the enzymatic hydrolyses of Isomaltoheptaose and isomaltose than those observed for the wild-type enzyme. This is the reason that dextran glucosidase, whose loop 4 is shorter in length, shows broader substrate chain-length specificity than does SAM1606 alpha-glucosidase.
Mutational modulation of substrate bond-type specificity and thermostability of glucoamylase from Aspergillus awamori by replacement with short homologue active site sequences and thiol/disulfide engineering.[Pubmed:8679632]
Biochemistry. 1996 Jul 2;35(26):8696-704.
Rational protein engineering based on three-dimensional structure, sequence alignment, and previous mutational analysis served to increase thermostability and modulate bond-type specificity in glucoamylase from Aspergillus awamori. The single free cysteine, Cys320, became disulfide bonded in the Ala246 --> Cys mutant, thus enhancing T50 by 4 degrees C to 73 degrees C. Compared to wild-type, Ala246 --> Cys was roughly twice as active at 66 degrees C, but half as active at 45 degrees C. The alternative, elimination of the thiol group in Cys320 --> Ala, barely improved thermostability or altered activity. Secondly, to acquire exceptionally high specificity toward alpha-1,6 glucosidic linkages, characteristic of Hormoconis resinae glucoamylase, two short sequential mutants, Val181 --> Thr/Asn182 --> Tyr/Gly183 --> Ala(L3 glucoamylase) and Pro307 --> Ala/Thr310 --> Val/Tyr312 --> Met/Asn313 --> Gly (L5 glucoamylase), were made. These homologue mutants are located in the (alpha/alpha)6-fold of the catalytic domain in segments that connect alpha-helices 5 and 6 and alpha-helices 9 and 10, respectively. The kinetics of malto- and isomaltooligosaccharides hydrolysis clearly demonstrated that combination of the mutations in L3L5 compensated adverse effects of the single replacements in L3 or L5 glucoamylases to yield wild-type or higher activity. On alpha-1,4-linked substrates, typically Km increased 2-fold for L3, and Kcat decreased up to 15-fold for L5 glucoamylase. In contrast, on alpha-1,6-linked substrates L3 showed both a 2-fold increase in Km and a 3-fold decrease in kcat, while L5 GA caused a similar kcat reduction, but up to 9-fold increase in Km. L3L5 glucoamylase had remarkably low Km for isomaltotriose through Isomaltoheptaose and elevated kcat on isomaltose, resulting in an approximately 2-fold improved catalytic efficiency (kcat/Km). Rational loop replacement thus proved powerful in achieving variants with enhanced properties of a highly evolved enzyme.
Glycosylation of a VH residue of a monoclonal antibody against alpha (1----6) dextran increases its affinity for antigen.[Pubmed:2459288]
J Exp Med. 1988 Sep 1;168(3):1099-109.
We have observed that antidextran hybridomas with potential N-linked glycosylation sites in VH have higher affinity for polymeric dextran and for Isomaltoheptaose than those lacking potential glycosylation sites. In these studies we have used gene transfection and expression techniques to verify that the carbohydrate addition sites in VH were used. The carbohydrate of the VH region was accessible for binding by the lectin Con A. By ELISA analysis it was demonstrated that the aKa of the antibody for dextran was influenced by the presence of carbohydrate in VH, with the aglycosylated antibody having an aKa 15-fold lower than its untreated counterpart. The aKa for antigen of antibodies that contain carbohydrate only in their constant region was unaffected by lack of carbohydrate. Thus, not only the amino acid sequence of the variable region but also its carbohydrate moieties can determine the magnitude of the antigen-antibody interaction.
Immunochemical studies on monoclonal antibodies to stearyl-isomaltotetraose from C58/J and a C57BL/10 nude mouse.[Pubmed:2443831]
Mol Immunol. 1987 Apr;24(4):333-8.
Six hybridomas, five from C58/J and one from C57BL/10 nu/nu, immunized with stearyl-isomaltotetraose (S-IM4) were established. One produced IgG3, one IgM and four IgA. The specificities and sizes of the antibody combining sites were determined by quantitative precipitin and ELISA quantitative inhibition assays. All cross-react with alpha(1----6)dextran B512. Their combining sites were complementary to five to seven glucose residues. Association constants of hybridoma antibodies for dextran B512 ranged from 10(3) to 10(5) ml/g, and for Isomaltoheptaose (IM7) from 10(3) to 10(4)M-1. Two hybridoma antibodies, one IgA and another IgM derived from different fusions have identical inhibition curves and combining sites, and very close association constants. It will be important to establish whether they have very similar or identical nucleotide sequences in their variable regions. These studies provide further insight into the specificity and repertoire of antidextran combining sites.
Oligosaccharide alterations of rat glioblast membrane-bound glycoproteins during differentiation induced by glia maturation factor.[Pubmed:20493026]
Neurochem Int. 1986;8(1):31-40.
Differentiation of normal glioblasts was induced by glia maturation factor (GMF), and the structural change in the oligosaccharide chains of the plasmalemmal glycoproteins was investigated. After the glycopeptides obtained by trypsin treatment of the intact cells had been digested with pronase, the resulting glycopeptides were separated into 4 fractions by gel filtration. The first 2 fractions were found to contain mainly N-glycosidically linked glycopeptides, and the last 2, O-linked oligosaccharides. There were a variety of N-linked oligosaccharides whose apparent molecular weights were greater than that of Isomaltoheptaose. As compared to those, O-linked oligosaccharides were fewer in type and lower in molecular weight. The N-linked oligosaccharides corresponding to isomaltohepta- decaose and larger saccharide chains augmented in differentiated glioblasts, whereas the N-linked oligosaccharides smaller than isomaltoheptade- caose decreased. The turnover rate of the high molecular weight oligosaccharides was faster than that of other membrane oligosaccharides, and was accelerated by GMF treatment. The content of an O-linked oligosaccharide fraction increased after GMF treatment.
Immunochemical studies of conjugates of isomaltosyl oligosaccharides to lipid: production and characterization of mouse hybridoma antibodies specific for stearyl-isomaltosyl oligosaccharides.[Pubmed:2415810]
Mol Immunol. 1985 Sep;22(9):1021-37.
Twelve C57BL/6J hybridoma clones, 9, 2 and 1 from mice immunized with stearyl-isomaltotetraose, stearyl-isomaltopentaose and stearyl-isomaltohexaose respectively were characterized. Seven produced IgA and 5 IgM. The specificities and sizes of their combining sites were determined by quantitative precipitin and precipitin inhibition assays. All 12 hybridoma antibodies precipitated with alpha 1----6 dextran B512 and linear dextran LD7, indicating that they recognize an internal -Glc alpha 1----6Glc alpha 1----6Glc- determinant. This in contrast with the results with rabbit antisera obtained in response to the same immunogen which recognize the non-reducing terminal determinant Glc alpha 1----6Glc alpha 1----6Glc-. Of the 12 hybridoma antibodies, 1 has an antibody combining site complementary to 4 alpha 1----6-linked glucoses while others have combining sites complementary to isomaltohexaose or Isomaltoheptaose. The large combining-site sizes found in C57BL/6 hybridoma clones may be related to the pre-existing clonal repertoire in this strain. Binding constants of monomers of these antibodies for dextran B512 and Isomaltoheptaose determined by affinity electrophoresis range from 1.4 X 10(3) to 4.6 X 10(5) ml/g and from 1.2 X 10(3) to 3.5 X 10(4) M-1 respectively, which is consistent with previous studies in the anti-dextran B512 system. The use of synthetic glycolipids as antigens enables us to study the gene control of antibody responses to glycolipids and to investigate the combining-site specificities of antibodies to a single antigenic determinant. Results so far show that all 12 hybridoma proteins are different despite the simplicity of the antigens. The findings provide further insight into the specificity of antibody combining sites.
Association constants of hybridoma antibodies specific for alpha (1 leads to 6) linked dextran determined by affinity electrophoresis.[Pubmed:6178962]
Mol Immunol. 1982 Mar;19(3):389-97.
Binding constants of monomers of seven BALB/c IgM, four BALB/c IgA, and one C57BL/6 IgA anti-alpha (1 leads to 6) dextran hybridoma antibodies with dextran B512 and with Isomaltoheptaose were determined by affinity electrophoresis. Bindings constants to dextran range from 1.52 X 10(5) to 4.43 X 10(5) ml/g for the five IgA monomers and from 1.70 X 10(3) to 6.10 X 10(4) ml/g for the seven IgM monomers. Antibody monomers containing both specific and nonspecific (derived from the myeloma cell that was used to generate the hybridomas) light chains are shown to have association constants with dextran 6 to 30-fold lower than monomers containing only specific light chain, suggesting that the association of specific heavy chain with nonspecific light chain does not result in an anti-dextran combining site. Binding constants with Isomaltoheptaose range from 1.45 X 10(4) to 7.01 X 10(4)/M for the IgA proteins and from 6.46 X 10(3) to 7.70 X 10(4)/M for the IgM proteins. The binding constants with dextran and with Isomaltoheptaose, and the electrophoretic, immunochemical and idiotypic characteristics of the hybridoma proteins are discussed.
Binding constants of NZB myeloma antidextrans for dextrans and isomaltose oligosaccharides determined by affinity electrophoresis.[Pubmed:469244]
J Immunol. 1979 Sep;123(3):1162-8.
Association constants of dextrans (Ka) and oligosaccharides (Kia) from NZB myeloma antidextrans (PC3858 and PC3936) were studied by affinity electrophoresis. With linear dextrans or with those with a low degree of branching, Ka ranged from 2.7 X 10(3) to 5.4 X 10(4) ml/g for PC3858 and from 1.3 X 10(4) to 2.6 X 10(5) ml/g for PC3936. Completely linear alpha-(1 leads to 6)-linked dextrans, LD7 and D3, showed relatively high affinities for the two NZB antidextrans. With oligosaccharides, the Kia value increased as the number oa alpha-(1 leads to 6)-linked glycosyl residues increased. Isomaltoheptaose (IM7) showed the highest Kia (1.9 X 10(4) M-1 for PC3858 and 1.63 X 10(4) M-1 for PC3936), whereas isomaltose (IM2) had the lowest Kia (2.36 X 10(2)M-1 for PC3858 and 1.32 X 10(2)M-1 for PC3936). Pullulan and glycogen showed very weak affinity for PC3936, but they did not react at all with PC3858. These findings indicate that NZB myeloma antidextrans, PC3858 and PC3936, are specific for internal chains of alpha-(1 leads to 6)-linked dextrans. Data on the precision with which Ka and Kia can be determined are presented.


