Inactivation of genes encoding redox enzymes leads to structural diversification of bioactive atypical angucyclines
A research group from the South China Sea Institute of Oceanology of the Chinese Academy of Sciences has reported the diversification of the structure of bioactive atypical angucyclines through the inactivation of genes encoding the redox enzyme in fluostatin biosynthesis.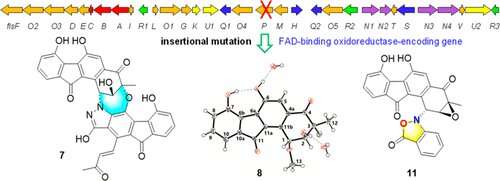
The study was published in the Journal of Natural Products on August 12.
Atypical angucyclines are a class of important natural products with antibacterial, antitumor, and enzyme inhibitory activities. Biosynthetic redox enzymes often play an important role in the structural richness of natural products.
In the biosynthetic gene clusters (BGCs) of atypical angucyclines, a number of redox enzymes have been found, but their biosynthetic functions are still unclear.
In this study, researchers performed in vivo gene disruption of the flsP gene encoding a redox enzyme in the fluostatin gene cluster, and reported the structural diversification of fluostatin-related atypical angucyclines. Bioinformatic analysis showed that the oxidoreductase encoding flsP was likely involved in the modification of the highly oxygenated A-ring of fluostatins.
A subsequent insertional deletion of flsP in Micromonospora rosaria SCSIO N160, the fluostatin producer, led to the discovery of a series of atypical angucycline derivatives.
In their previous study published in The Journal of Organic Chemistry, four new angucyclinone derivatives were discovered by inactivating the flsO1 gene encoding a flavoenzyme. In particular, it was revealed that FlsO1 was a benzofluorene hydroxylase functionally equivalent to its homologous enzyme AlpK in kinamycin biosynthesis.


