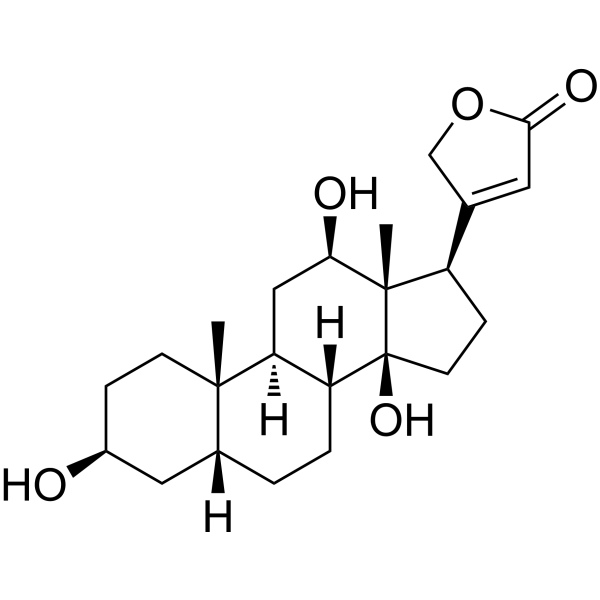
DigoxigeninCAS# 1672-46-4 |
Quality Control & MSDS
Package In Stock
Number of papers citing our products

| Cas No. | 1672-46-4 | SDF | File under preparation. |
| PubChem ID | N/A | Appearance | Powder |
| Formula | C23H34O5 | M.Wt | 390.51 |
| Type of Compound | Cardenolides and its Sapogenins | Storage | Desiccate at -20°C |
| Synonyms | 101 claimed sequence,Lanadigenin,3β-Digoxigenin | ||
| Solubility | Soluble in Chloroform,Dichloromethane,Ethyl Acetate,DMSO,Acetone,etc. | ||
| General tips | For obtaining a higher solubility , please warm the tube at 37 ℃ and shake it in the ultrasonic bath for a while.Stock solution can be stored below -20℃ for several months. We recommend that you prepare and use the solution on the same day. However, if the test schedule requires, the stock solutions can be prepared in advance, and the stock solution must be sealed and stored below -20℃. In general, the stock solution can be kept for several months. Before use, we recommend that you leave the vial at room temperature for at least an hour before opening it. |
||
| About Packaging | 1. The packaging of the product may be reversed during transportation, cause the high purity compounds to adhere to the neck or cap of the vial.Take the vail out of its packaging and shake gently until the compounds fall to the bottom of the vial. 2. For liquid products, please centrifuge at 500xg to gather the liquid to the bottom of the vial. 3. Try to avoid loss or contamination during the experiment. |
||
| Shipping Condition | Packaging according to customer requirements(5mg, 10mg, 20mg and more). Ship via FedEx, DHL, UPS, EMS or other couriers with RT, or blue ice upon request. | ||

Digoxigenin Dilution Calculator

Digoxigenin Molarity Calculator
| 1 mg | 5 mg | 10 mg | 20 mg | 25 mg | |
| 1 mM | 2.5608 mL | 12.8038 mL | 25.6075 mL | 51.2151 mL | 64.0188 mL |
| 5 mM | 0.5122 mL | 2.5608 mL | 5.1215 mL | 10.243 mL | 12.8038 mL |
| 10 mM | 0.2561 mL | 1.2804 mL | 2.5608 mL | 5.1215 mL | 6.4019 mL |
| 50 mM | 0.0512 mL | 0.2561 mL | 0.5122 mL | 1.0243 mL | 1.2804 mL |
| 100 mM | 0.0256 mL | 0.128 mL | 0.2561 mL | 0.5122 mL | 0.6402 mL |
| * Note: If you are in the process of experiment, it's necessary to make the dilution ratios of the samples. The dilution data above is only for reference. Normally, it's can get a better solubility within lower of Concentrations. | |||||

Calcutta University

University of Minnesota

University of Maryland School of Medicine

University of Illinois at Chicago

The Ohio State University

University of Zurich

Harvard University

Colorado State University

Auburn University

Yale University

Worcester Polytechnic Institute

Washington State University

Stanford University

University of Leipzig

Universidade da Beira Interior

The Institute of Cancer Research

Heidelberg University

University of Amsterdam

University of Auckland

TsingHua University

The University of Michigan

Miami University

DRURY University

Jilin University

Fudan University

Wuhan University

Sun Yat-sen University

Universite de Paris

Deemed University

Auckland University

The University of Tokyo

Korea University
- Dichlorogelignate
Catalog No.:BCX1888
CAS No.:164030-91-5
- Lasiokaurinin
Catalog No.:BCX1887
CAS No.:52554-74-2
- 3-Epigitoxigenin
Catalog No.:BCX1886
CAS No.:465-10-1
- Sibiriquinone A
Catalog No.:BCX1885
CAS No.:723300-08-1
- 2,4,6-Trimethoxyl-3-methylacetophenone
Catalog No.:BCX1884
CAS No.:39701-13-8
- Dehydrodanshenol A
Catalog No.:BCX1883
CAS No.:1444618-61-4
- Hymenoxin
Catalog No.:BCX1882
CAS No.:56003-01-1
- Pebrellin
Catalog No.:BCX1881
CAS No.:13509-93-8
- Chrysindin A
Catalog No.:BCX1880
CAS No.:1374852-85-3
- 3-Epidigitoxigenin
Catalog No.:BCX1879
CAS No.:545-52-8
- Marsdenoside B
Catalog No.:BCX1878
CAS No.:858360-57-3
- 11α-O-Tigloyl-12β-O-acetyltenacigenin B
Catalog No.:BCX1877
CAS No.:154022-51-2
- 17α-Gitoxigenin
Catalog No.:BCX1890
CAS No.:4433-59-4
- Digitalin, 16-anhydro-
Catalog No.:BCX1891
CAS No.:7044-34-0
- Abrine D
Catalog No.:BCX1896
CAS No.:862504-05-0
- 4-((2-ethylidene-4-hydroxy-6-methylhept-5-en-1-yl)oxy)-bergaptol
Catalog No.:BCX1892
CAS No.:2519548-06-0
- 3,5,6,6-tetramethyl-4,5,5a,6,7,8-hexahydrocyclopenta[c]pentalen-2(1H)-one
Catalog No.:BCX1893
CAS No.:1689570-10-2
- Genistein 7-O-β-D-glucopyranoside-4’-O-[α-L-rhamnopyranosyl-(1→2)-β-D-glucopyranoside]
Catalog No.:BCX2014
CAS No.:361447-29-2
- 3-epi-Tilifodiolide
Catalog No.:BCX1897
CAS No.:1983982-42-8
- Licorisoflavan I
Catalog No.:BCX1894
CAS No.:2444736-45-0
- Tenuifoliose D
Catalog No.:BCX1895
CAS No.:139682-04-5
- Ipalbinium
Catalog No.:BCX1898
CAS No.:110200-24-3
- Isolithospermic acid
Catalog No.:BCX1899
CAS No.:1217879-76-9
- Digitoxigenin monodigitoxoside
Catalog No.:BCX1990
CAS No.:18404-43-8
Carbon nanoparticle-based lateral flow assay for the detection of specific double-tagged DNA amplicons of Paracoccidioides spp.[Pubmed:38671236]
Mikrochim Acta. 2024 Apr 26;191(5):287.
To overcome the limitations of current methods for diagnosing paracoccidioidomycosis (PCM), it is critical to develop novel diagnostic strategies that can be implemented in low-resource settings and dramatically improve turnaround times. This study focused on the development of a portable molecular test to screen for Paracoccidioides spp. The proposed approach integrated double-tagging polymerase chain reaction (PCR) and a paper-based lateral flow assay (LFA) for readout, using carbon nanoparticles as a signal generation system. Primers tagged with biotin and Digoxigenin were employed to conduct the double-tagging PCR, which can be conveniently carried out on portable thermocyclers. This method can generate billions of tagged DNA copies from a single target molecule, which can be rapidly detected by the LFA platform, providing results within minutes. Avidin-modified carbon nanoparticles served as a signal generation system, enabling detection in the immunochromatographic assay. The LFA demonstrated the capability to detect double-tagged amplicons as low as 0.21 ng or 0.10 ng, depending on whether the results were assessed visually or with a smartphone equipped with an image processor. These findings suggest that the proposed approach holds great promise as a point-of-care diagnostic tool for the early and accurate detection of PCM in low-resource settings. The diagnostic test is rapid and inexpensive, requires minimal handling and can be easily introduced into the general practitioner's armoury for ambulatory screening of infection. This innovative approach has the potential to make a substantial contribution to PCM diagnosis, ultimately reducing morbidity and mortality associated with this disease.
Development of nucleic acid lateral flow immunoassay for molecular detection of Entamoeba moshkovskii and Entamoeba dispar in stool samples.[Pubmed:38503871]
Sci Rep. 2024 Mar 19;14(1):6635.
Entamoeba moshkovskii, recently known as a possible pathogenic amoeba, and the non-pathogenic Entamoeba dispar are morphologically indistinguishable by microscopy. Although PCR was used for differential diagnosis, gel electrophoresis is labor-intensive, time-consuming, and exposed to hazardous elements. In this study, nucleic acid lateral flow immunoassay (NALFIA) was developed to detect E. moshkovskii and E. dispar by post-PCR amplicon analysis. E. moshkovskii primers were labeled with Digoxigenin and biotin whereas primers of E. dispar were lebeled with FITC and Digoxigenin. The gold nanoparticles were labeled with antibodies corresponding to particular labeling. Based on the established assay, NALFIA could detect as low as 975 fg of E. moshkovskii target DNA (982 parasites or 196 parasites/microliter), and 487.5 fg of E. dispar target DNA (444 parasites or 89 parasites/microliter) without cross-reactivity to other tested intestinal organisms. After testing 91 stool samples, NALFIA was able to detect seven E. moshkovskii (87.5% sensitivity and 100% specificity) and eight E. dispar samples (66.7% sensitivity and 100% specificity) compared to real-time PCR. Interestingly, it detected three mixed infections as real-time PCR. Therefore, it can be a rapid, safe, and effective method for the detection of the emerging pathogens E. moshkovskii and E. dispar in stool samples.
Single-molecule tethering methods for membrane proteins.[Pubmed:38492954]
Methods Enzymol. 2024;694:263-284.
Molecular tethering of a single membrane protein between the glass surface and a magnetic bead is essential for studying the structural dynamics of membrane proteins using magnetic tweezers. However, the force-induced bond breakage of the widely-used Digoxigenin-antiDigoxigenin tether complex has imposed limitations on its stable observation. In this chapter, we describe the procedures of constructing highly stable single-molecule tethering methods for membrane proteins. These methods are established using dibenzocyclooctyne click chemistry, traptavidin-biotin binding, SpyCatcher-SpyTag conjugation, and SnoopCatcher-SnoopTag conjugation. The molecular tethering approaches allow for more stable observation of structural transitions in membrane proteins under force.
Generalized biomolecular modeling and design with RoseTTAFold All-Atom.[Pubmed:38452047]
Science. 2024 Apr 19;384(6693):eadl2528.
Deep-learning methods have revolutionized protein structure prediction and design but are presently limited to protein-only systems. We describe RoseTTAFold All-Atom (RFAA), which combines a residue-based representation of amino acids and DNA bases with an atomic representation of all other groups to model assemblies that contain proteins, nucleic acids, small molecules, metals, and covalent modifications, given their sequences and chemical structures. By fine-tuning on denoising tasks, we developed RFdiffusion All-Atom (RFdiffusionAA), which builds protein structures around small molecules. Starting from random distributions of amino acid residues surrounding target small molecules, we designed and experimentally validated, through crystallography and binding measurements, proteins that bind the cardiac disease therapeutic Digoxigenin, the enzymatic cofactor heme, and the light-harvesting molecule bilin.
TUNEL-n-DIFL Method for Detection and Estimation of Apoptosis Specifically in Neurons and Glial Cells in Mixed Culture and Animal Models of Central Nervous System Diseases and Injuries.[Pubmed:38427225]
Methods Mol Biol. 2024;2761:1-26.
Detection of merely apoptosis does not reveal the type of central nervous system (CNS) cells that are dying in the CNS diseases and injuries. In situ detection and estimation of amount of apoptosis specifically in neurons or glial cells (astrocytes, oligodendrocytes, and microglia) can unveil valuable information for designing therapeutics for protection of the CNS cells and functional recovery. A method was first developed and reported from our laboratory for in situ detection and estimation of amount of apoptosis precisely in neurons and glial cells using in vitro and in vivo models of CNS diseases and injuries. This is a combination of terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) and double immunofluorescent labeling (DIFL) or simply TUNEL-n-DIFL method for in situ detection and estimation of amount of apoptosis in a specific CNS cell type. An anti-Digoxigenin (DIG) IgG antibody conjugated with 7-amino-4-methylcoumarin-3-acetic acid (AMCA) for blue fluorescence, fluorescein isothiocyanate (FITC) for green fluorescence, or Texas Red (TR) for red fluorescence can be used for in situ detection of apoptotic cell DNA, which is earlier labeled with TUNEL using alkali-stable DIG-11-dUTP. A primary anti-NeuN (neurons), anti-GFAP (astrocytes), anti-MBP (oligodendrocytes), or anti-OX-42 (microglia) IgG antibody and a secondary IgG antibody conjugated with one of the above fluorophores (other than that of ani-DIG antibody) are used for in situ detection of apoptosis in a specific CNS cell type in the mixed culture and animal models of the CNS diseases and injuries.
Addressing Current Challenges in Poultry Meat Safety: Development of a Cultivation and Colony Hybridization Approach to Recover Enterotoxigenic Clostridium perfringens from Broiler Chicken Carcasses.[Pubmed:38251337]
Pathogens. 2023 Dec 28;13(1):30.
Enterotoxigenic Clostridium perfringens is one of the main causes of foodborne illness in Canada. The use of a conventional bacterial culture approach to isolate enterotoxigenic C. perfringens from poultry meat is common. This approach is based on the phenotype attributable to a double hemolysis phenomenon, whereas few enterotoxigenic strains of C. perfringens produce it, which further complicates the study of the reservoirs of this important pathogen. The objectives of the current study were to validate the ability of a Digoxigenin-labeled probe to detect the C. perfringens cpe gene and to validate the use of either a filtration or a direct plating approach, combined with colony hybridization to detect enterotoxigenic C. perfringens. Pure DNA and pure colonies of enterotoxigenic C. perfringens and broiler chicken carcass rinsate samples were subjected to colony hybridization. The results showed that the synthesized DNA probe can detect the cpe gene from both DNA and pure colonies of enterotoxigenic C. perfringens, and from colonies grown from carcass rinsates artificially contaminated with enterotoxigenic C. perfringens. Our study suggests that this isolation method is a promising tool for a better understanding of the epidemiology of this zoonotic pathogen.
Centrifugal microfluidic system for colorimetric sample-to-answer detection of viral pathogens.[Pubmed:38226743]
Lab Chip. 2024 Feb 13;24(4):668-679.
We describe a microfluidic system for conducting thermal lysis, polymerase chain reaction (PCR) amplification, hybridization, and colorimetric detection of foodborne viral organisms in a sample-to-answer format. The on-chip protocol entails 24 steps which are conducted by a centrifugal platform that allows for actuating liquids pneumatically during rotation and so facilitates automation of the workflow. The microfluidic cartridge is fabricated from transparent thermoplastic polymers and accommodates assay components along with an embedded micropillar array for detection and read-out. A panel of oligonucleotide primers and probes has been developed to perform PCR and hybridization assays that allows for identification of five different viruses, including pathogens such as norovirus and hepatitis A virus (HAV) in a multiplexed format using Digoxigenin-labelled amplicons and immunoenzymatic conversion of a chromogenic substrate. Using endpoint detection, we demonstrate that the system can accurately and repetitively (n = 3) discriminate positive and negative signals for HAV at 350 genome copies per muL. As part of the characterization and optimization process, we show that the implementation of multiple (e.g., seven) micropillar arrays in a narrow fluidic pathway can lead to variation (up to 50% or more) in the distribution of colorimetric signal deriving from the assay. Numerical modeling of flow behaviour was used to substantiate these findings. The technology-by virtue of automation-can provide a pathway toward rapid detection of viral pathogens, shortening response time in food safety surveillance, compliance, and enforcement as well as outbreak investigations.
Examining Cell-Specific Localization in Aedes aegypti Tissues with Fluorescence In Situ Hybridization.[Pubmed:38087467]
Cold Spring Harb Protoc. 2023 Dec 8.
Fluorescence in situ hybridization (FISH) is a macromolecular recognition tool that uses RNA or DNA fragments combined with fluorophore- or Digoxigenin-coupled nucleotides as probes to examine transcript localization through the presence or absence of complementary sequences in fixed tissues or samples under a fluorescent microscope. FISH technology has been highly effective for mapping genes and constructing a visual map of animal genomes. Here, we describe the application of FISH technology in the Aedes aegypti mosquito, where it is specifically used to localize receptor transcripts in gut tissues/organs. The methods presented highlight the synthesis of RNA probes and describe the 2-d process of incubating the tissues/organs with the RNA probes. We also describe tyramide signal amplification for improved signal detection.
A highly sensitive biotin-based probe for small RNA northern blot and its application in dissecting miRNA function in pepper.[Pubmed:38078656]
Plant J. 2024 Apr;118(1):263-276.
Small RNAs play important roles in regulation of plant development and response to various stresses. Northern blot is an important technique in small RNA research. Isotope- and biotin- (or Digoxigenin) labeled probes are frequently used in small RNA northern blot. However, isotope-based probe is limited by strict environmental regulation and availability in many places in the world while biotin-based probe is usually suffered from low sensitivity. In this study, we developed a T4 DNA polymerase-based method for incorporation of a cluster of 33 biotin-labeled C in small RNA probe (T4BC33 probe). T4BC33 probe reaches similar sensitivity as (32)P-labeled probe in dot blot and small RNA northern blot experiments. Addition of locked nucleic acids in T4BC33 probe further enhanced its sensitivity in detecting low-abundance miRNAs. With newly developed northern blot method, expression of miR6027 and miR6149 family members was validated. Northern blot analysis also confirmed the successful application of virus-based miRNA silencing in pepper, knocking down accumulation of Can-miR6027a and Can-miR6149L. Importantly, further analysis showed that knocking-down Can-miR6027a led to upregulation of a nucleotide binding-leucine rich repeat domain protein coding gene (CaRLb1) and increased immunity against Phytophthora capsici in pepper leaves. Our study provided a highly sensitive and convenient method for sRNA research and identified new targets for genetic improvement of pepper immunity against P. capsici.
Application of N-Terminal Site-Specific Biotin and Digoxigenin Conjugates to Clinical Anti-drug Antibody Assay Development.[Pubmed:38050929]
Bioconjug Chem. 2024 Feb 21;35(2):174-186.
Biotin- and Digoxigenin (DIG)-conjugated therapeutic drugs are critical reagents used for the development of anti-drug antibody (ADA) assays for the assessment of immunogenicity. The current practice of generating biotin and DIG conjugates is to label a therapeutic antibody with biotin or DIG via primary amine groups on lysine or N-terminal residues. This approach modifies lysine residues nonselectively, which can impact the ability of an ADA assay to detect those ADAs that recognize epitopes located at or near the modified lysine residue(s). The impact of the lysine modification is considered greater for therapeutic antibodies that have a limited number of lysine residues, such as the variable heavy domain of heavy chain (VHH) antibodies. In this paper, for the first time, we report the application of site-specifically conjugated biotin- and DIG-VHH reagents to clinical ADA assay development using a model molecule, VHHA. The site-specific conjugation of biotin or DIG to VHHA was achieved by using an optimized reductive alkylation approach, which enabled the majority of VHHA molecules labeled with biotin or DIG at the desirable N-terminus, thereby minimizing modification of the protein after labeling and reducing the possibility of missing detection of ADAs. Head-to-head comparison of biophysical characterization data revealed that the site-specific biotin and DIG conjugates demonstrated overall superior quality to biotin- and DIG-VHHA prepared using the conventional amine coupling method, and the performance of the ADA assay developed using site-specific biotin and DIG conjugates met all acceptance criteria. The approach described here can be applied to the production of other therapeutic-protein- or antibody-based critical reagents that are used to support ligand binding assays.
Expression of secretory calcium-binding phosphoprotein (scpp) genes in medaka during the formation and replacement of pharyngeal teeth.[Pubmed:37821862]
BMC Oral Health. 2023 Oct 11;23(1):744.
BACKGROUND: Analyses of tooth families and tooth-forming units in medaka with regard to tooth replacement cycles and the localization of odontogenic stem cell niches in the pharyngeal dentition clearly indicate that continuous tooth replacement is maintained. The secretory calcium-binding phosphoprotein (scpp) gene cluster is involved in the formation of mineralized tissues, such as dental and bone tissues, and the genes encoding multiple SCPPs are conserved in fish, amphibians, reptiles, and mammals. In the present study, we examined the expression patterns of several scpp genes in the pharyngeal teeth of medaka to elucidate their roles during tooth formation and replacement. METHODS: Himedaka (Japanese medaka, Oryzias latipes) of both sexes (body length: 28 to 33 mm) were used in this study. Real-time quantitative reverse transcription-polymerase chain reaction (PCR) (qPCR) data were evaluated using one-way analysis of variance for multi-group comparisons, and the significance of differences was determined by Tukey's comparison test. The expression of scpp genes was examined using in situ hybridization (ISH) with a Digoxigenin-labeled, single-stranded antisense probe. RESULTS: qPCR results showed that several scpp genes were strongly expressed in pharyngeal tissues. ISH analysis revealed specific expression of scpp1, scpp5, and sparc in tooth germ, and scpp5 was continually expressed in the odontoblasts of teeth attached to pedicles, but not in the osteoblasts of pedicles. In addition, many scpp genes were expressed in inner dental epithelium (ide), but not in odontoblasts, and scpp2 consistently showed epithelial-specific expression in the functional teeth. Taken together, these data indicate that specific expression of scpp2 and scpp5 may play a critical role in pharyngeal tooth formation in medaka. CONCLUSION: We characterized changes in the expression patterns of scpp genes in medaka during the formation and replacement of pharyngeal teeth.
Decorated DNA-Based Scaffolds as Lateral Flow Biosensors.[Pubmed:37804080]
Angew Chem Int Ed Engl. 2023 Nov 20;62(47):e202313243.
Here we develop Lateral Flow Assays (LFAs) that employ as functional elements DNA-based structures decorated with reporter tags and recognition elements. We have rationally re-engineered tile-based DNA tubular structures that can act as scaffolds and can be decorated with recognition elements of different nature (i.e. antigens, aptamers or proteins) and with orthogonal fluorescent dyes. As a proof-of-principle we have developed sandwich and competitive multiplex lateral flow platforms for the detection of several targets, ranging from small molecules (Digoxigenin, Dig and dinitrophenol, DNP), to antibodies (Anti-Dig, Anti-DNP and Anti-MUC1/EGFR bispecific antibodies) and proteins (thrombin). Coupling the advantages of functional DNA-based scaffolds together with the simplicity of LFAs, our approach offers the opportunity to detect a wide range of targets with nanomolar sensitivity and high specificity.
Immunohistochemical Detection of the Expressed BRCA1 and BRCA2 Proteins in Microenvironment of Malignant Breast Cancerous Tissues Infected with Human Mammary Tumor Virus.[Pubmed:37774080]
Asian Pac J Cancer Prev. 2023 Sep 1;24(9):3261-3267.
OBJECTIVE: The objective of the study was to examine the prevalence of HMTV infection, its associations with breast malignant tissues, and the expression of BRCA1 and BRCA2 proteins. METHODS: One hundred archival breast tissues, 40 biopsies from female patients with breast cancer (BC), and 20 healthy breast tissues from the control group were used in the study. Immunohistochemical analysis was conducted to detect the expressed BRCA1 and BRCA2 proteins. Digoxigenin-labeled HMTV probes were used in chromogenic in situ hybridization for the identification of HMTV in breast tumor tissues. The complementary sequence sites of the HMTV probe sequences were stained by NBT/BCIP as blue signals. RESULTS: There were 12 out of 40 (30%) benign breast tumorous tissues and 14 out of 40 (35%) BC tissues, while healthy control breast tissues were 10% (2 out of 20 tissues). Positive immunohistochemical (IHC) reactions for BRCA2 protein were observed in 12 out of 40 BC tissues (30.0%), 25% of benign breast tumorous tissues, and 5% of the control group. A significant (p < 0.05) statistical difference in the percentages of HMTV in the studied groups was found. CONCLUSION: HMTV might contribute to the development of subsets of benign and malignant breast tumors. The observed rates of defective or mutated BRCA1 and BRCA2 genes in healthy tissues indicate a role in the development of breast tumors.
Fluorescence in situ Localization of Pri-miRNAs in Isolated Arabidopsis thaliana Nuclei.[Pubmed:37753471]
Bio Protoc. 2023 Sep 20;13(18):e4824.
Here, we present an approach combining fluorescence in situ hybridization (FISH) and immunolabeling for localization of pri-miRNAs in isolated nuclei of A. thaliana. The presented method utilizes specific DNA oligonucleotide probes, modified by addition of Digoxigenin-labeled deoxynucleotides to its 3' hydroxyl terminus by terminal deoxynucleotidyl transferase (TdT). The probes are then detected by immunolabeling of Digoxigenin (DIG) using specific fluorescent-labeled antibodies to visualize hybridized probes. Recently, we have applied this method to localize pri-miRNA156a, pri-miRNA163, pri-miRNA393a, and pri-miRNA414 in the nuclei isolated from leaves of 4-week-old A. thaliana. The present approach can be easily implemented to analyze nuclear distribution of diverse RNA classes, including mRNAs and pri-miRNAs in isolated fixed cells or nuclei from plant.
An antibody-based molecular switch for continuous small-molecule biosensing.[Pubmed:37738337]
Sci Adv. 2023 Sep 22;9(38):eadh4978.
We present a generalizable approach for designing biosensors that can continuously detect small-molecule biomarkers in real time and without sample preparation. This is achieved by converting existing antibodies into target-responsive "antibody-switches" that enable continuous optical biosensing. To engineer these switches, antibodies are linked to a molecular competitor through a DNA scaffold, such that competitive target binding induces scaffold switching and fluorescent signaling of changing target concentrations. As a demonstration, we designed antibody-switches that achieve rapid, sample preparation-free sensing of Digoxigenin and cortisol in undiluted plasma. We showed that, by substituting the molecular competitor, we can further modulate the sensitivity of our cortisol switch to achieve detection at concentrations spanning 3.3 nanomolar to 3.3 millimolar. Last, we integrated this switch with a fiber optic sensor to achieve continuous sensing of cortisol in a buffer and blood with <5-min time resolution. We believe that this modular sensor design can enable continuous biosensor development for many biomarkers.


