Cucumis sativus
Cucumis sativus
1. The products in our compound library are selected from thousands of unique natural products; 2. It has the characteristics of diverse structure, diverse sources and wide coverage of activities; 3. Provide information on the activity of products from major journals, patents and research reports around the world, providing theoretical direction and research basis for further research and screening; 4. Free combination according to the type, source, target and disease of natural product; 5. The compound powder is placed in a covered tube and then discharged into a 10 x 10 cryostat; 6. Transport in ice pack or dry ice pack. Please store it at -20 °C as soon as possible after receiving the product, and use it as soon as possible after opening.
Natural products/compounds from Cucumis sativus
- Cat.No. Product Name CAS Number COA
-
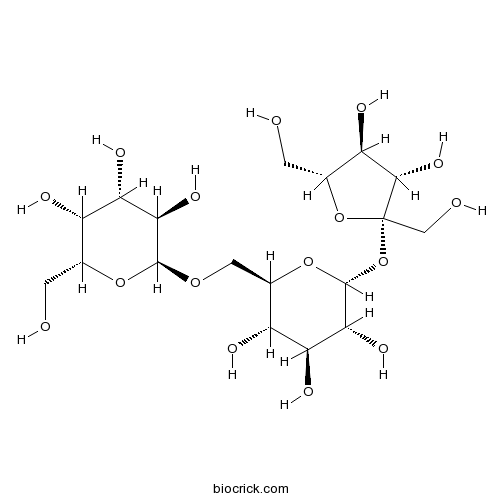
BCN8427
Raffinose512-69-6
Instructions
-
BCN3646
Dehydroeburicoic acid6879-05-6
Instructions

-
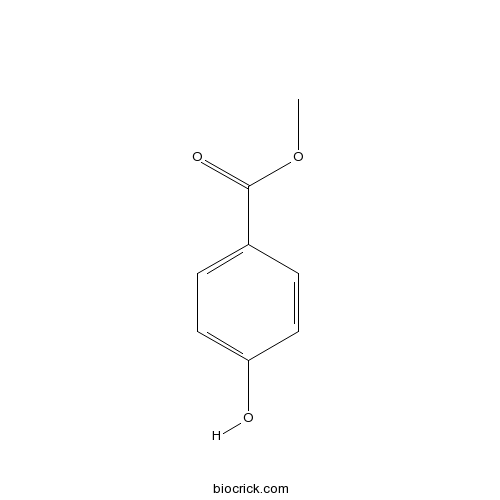
BCN4540
Methyl 4-hydroxybenzoate99-76-3
Instructions
-
BCN4546
4-Hydroxybenzoic acid99-96-7
Instructions

Prediction models for evaluating the uptake of heavy metals by cucumbers (Cucumis sativus L.) grown in agricultural soils amended with sewage sludge.[Pubmed: 30084016]
None
Comparative proteomic analysis of key proteins during abscisic acid-hydrogen peroxide-induced adventitious rooting in cucumber (Cucumis sativus L.) under drought stress.[Pubmed: 30082096]
None
Uptake, distribution and transformation of zerovalent iron nanoparticles in the edible plant Cucumis sativus.[Pubmed: 30078317]
Here, we investigated the fate of nanoscale zerovalent iron (nZVI) on the Cucumis sativus under both hydroponic and soil conditions. Seedlings were exposed to 0, 250 and 1000 mg/L (or mg/kg soil) nZVI during 6-9 weeks of a growth period. Ionic controls were prepared by using Fe-EDTA. None of the nZVI treatments affected the plant biomass. Based on the total iron contents and the superparamagnetic property of nZVI-exposed roots, there was no evidence of pristine nZVI translocation from the roots to shoots. Electron microscopy revealed that the transformed iron nanoparticles are stored in the root cell membrane and the vacuoles of the leaf parenchymal cells. X-ray absorption spectroscopy identified ferric citrate (41%) and iron (oxyhydr)oxides (59%) as the main transformed products in the roots. The shoot samples indicated a larger proportion of ferric citrate (60%) compared to iron (oxyhydr)oxides (40%). The 1.8-fold higher expression of the CsHA1 gene indicated that the plant-promoted transformation of nZVI was driven by protons released from the root layers. The current data provide a basis for two potential nZVI transformation pathways in Cucumis sativus: (1) interaction with low molecular weight organic acid ligands, and (2) dissolution-precipitation of the mineral products.
Cucumber and Tomato Volatiles: Influence on Attraction in the Melon Fly Zeugodacus cucurbitate (Diptera: Tephritidae).[Pubmed: 30041516]
The main hosts of the melon fly Zeugodacus cucurbitate are cultivated and wild cucurbitaceous plants. In eastern Africa, the melon fly is a major pest of the Solanaceae plant Solanum lycopersicum (tomato). We hypothesized that shared species-specific volatiles may play a role in host attraction. We tested this hypothesis by comparing the olfactory responses of the melon fly to Cucumis sativus (cucumber) (Cucurbitaceae) and tomato plant odors in behavioral and electrophysiological assays, followed by chemical analysis to identify the key compounds mediating the interactions. Our results identified 13 shared components between cucumber and tomato plant odors. A synthetic blend of seven of the shared components dominated by monoterpenes at concentrations mimicking the volatile bouquet of cucumber and tomato attracted both sexes of the melon fly. Our results suggest that the presence and quantity of specific compounds in host odors are the main predictors for host recognition in Z. cucurbitate.
STAYGREEN, STAY HEALTHY: a loss-of-susceptibility mutation in the STAYGREEN gene provides durable, broad-spectrum disease resistances for over 50 years of US cucumber production.[Pubmed: 30022503]
The Gy14 cucumber (Cucumis sativus) is resistant to oomyceteous downy mildew (DM), bacterial angular leaf spot (ALS) and fungal anthracnose (AR) pathogens, but the underlying molecular mechanisms are unknown. Quantitative trait locus (QTL) mapping for the disease resistances in Gy14 and further map-based cloning identified a candidate gene for the resistant loci, which was validated and functionally characterized by spatial-temporal gene expression profiling, allelic diversity and phylogenetic analysis, as well as local association studies. We showed that the triple-disease resistances in Gy14 were controlled by the cucumber STAYGREEN (CsSGR) gene. A single nucleotide polymorphism (SNP) in the coding region resulted in a nonsynonymous amino acid substitution in the CsSGR protein, and thus disease resistance. Genes in the chlorophyll degradation pathway showed differential expression between resistant and susceptible lines in response to pathogen inoculation. The causal SNP was significantly associated with disease resistances in natural and breeding populations. The resistance allele has undergone selection in cucumber breeding. The durable, broad-spectrum disease resistance is caused by a loss-of-susceptibility mutation of CsSGR. Probably, this is achieved through the inhibition of reactive oxygen species over-accumulation and phytotoxic catabolite over-buildup in the chlorophyll degradation pathway. The CsSGR-mediated host resistance represents a novel function of this highly conserved gene in plants.


