Jatropha curcas
Jatropha curcas
1. The products in our compound library are selected from thousands of unique natural products; 2. It has the characteristics of diverse structure, diverse sources and wide coverage of activities; 3. Provide information on the activity of products from major journals, patents and research reports around the world, providing theoretical direction and research basis for further research and screening; 4. Free combination according to the type, source, target and disease of natural product; 5. The compound powder is placed in a covered tube and then discharged into a 10 x 10 cryostat; 6. Transport in ice pack or dry ice pack. Please store it at -20 °C as soon as possible after receiving the product, and use it as soon as possible after opening.
Natural products/compounds from Jatropha curcas
- Cat.No. Product Name CAS Number COA
-
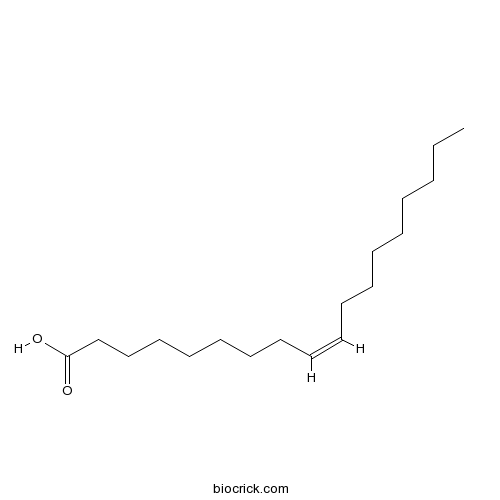
BCN7159
Oleic acid112-80-1
Instructions
-
BCN5433
Cimifugin37921-38-3
Instructions

-
BCN2502
Isovanillin621-59-0
Instructions

-
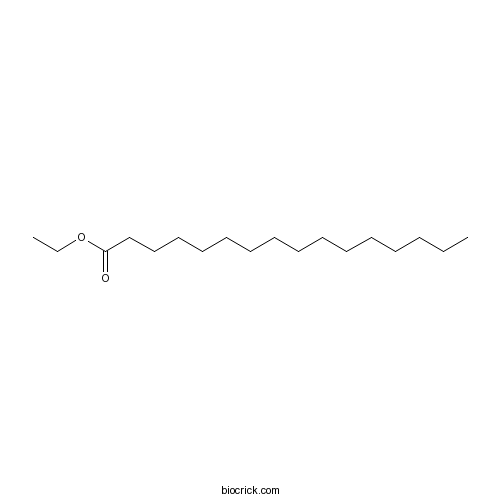
BCN8298
Palmitic acid ethyl ester628-97-7
Instructions
-
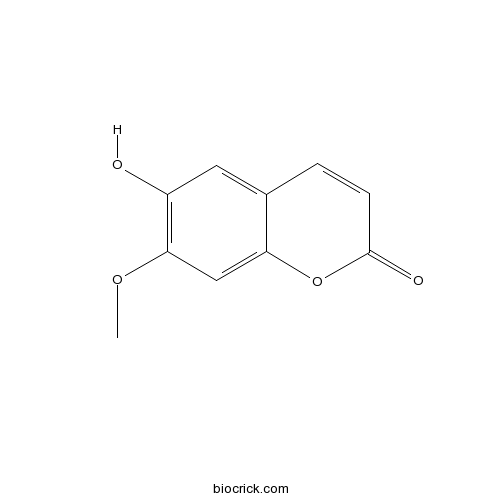
BCN4582
Isoscopoletin776-86-3
Instructions
Genome sequence of Jatropha curcas L., a non-edible biodiesel plant, provides a resource to improve seed-related traits.[Pubmed: 30059608]
Jatropha curcas (physic nut), a non-edible oilseed crop, represents one of the most promising alternative energy sources due to its high seed oil content, rapid growth, and adaptability to various environments. We report ~339 Mbp draft whole genome sequence of J. curcas var. Chai Nat using both the PacBio and Illumina sequencing platforms. We identified and categorized differentially expressed genes related to biosynthesis of lipid and toxic compound among four stages of seed development. Triacylglycerol (TAG), the major component of seed storage oil, is mainly synthesized by phospholipid:diacylglycerol acyltransferase in Jatropha, and continuous high expression of homologs of oleosin over seed development contributes to accumulation of high level of oil in kernels by preventing the breakdown of TAG. A physical cluster of genes for diterpenoid biosynthetic enzymes, including casbene synthases highly responsible for a toxic compound, phorbol ester, in seed cake, was syntenically highly conserved between Jatropha and castor bean. Transcriptomic analysis of female and male flowers revealed the up-regulation of a dozen family of TFs in female flower. Additionally, we constructed a robust species tree enabling estimation of divergence times among nine Jatropha species and five commercial crops in Malpighiales order. Our results will help researchers and breeders increase energy efficiency of this important oil seed crop by improving yield and oil content, and eliminating toxic compound in seed cake for animal feed. This article is protected by copyright. All rights reserved.
Jatropha half-sib family selection with high adaptability and genotypic stability.[Pubmed: 30001344]
Jatropha (Jatropha curcas) has become one of the most important species for producing biofuels. Currently, Genotype x Environment (GxE) interaction is the biggest challenge that breeders should solve to increase the section accuracy in the plant breeding. Therefore, the objectives in this study were to estimate the parameters in the 180 half-sib families in Jatropha evaluated for five production years, to verify the significance of the GxE interaction variance, to evaluate the adaptability and stability for each family based on three prediction methods, to select superior half-sib families based on the adaptability and stability analyses, and to predict the accuracy for the sixth production year. Jatropha half-sib families were classified and selected using the follow adaptability and stability methods: linear regression, bi-segmented linear regression and mixed models concepts called harmonic mean of the relative performance of genetic values (HMRPGV). The prediction accuracy was estimated by the Pearson correlation between the predicted genetic values by adaptability and stability methods and the phenotypic value in the sixth production year. In result, most half-sib families were classified as general adaptability and general stability for the evaluated traits. The selection gain obtained via HMRPGV was higher than other methods. The prediction accuracy for the sixth production year was 0.45. Therefore, HMRPGV is efficient to maximize the genetic gain, and it can be a useful strategy to select genotype with high adaptability and stability in Jatropha breeding as well as other species that should be evaluated for many years to take a suitable selection accuracy.
Specific enzyme functionalities of Fusarium oxysporum compared to host plants.[Pubmed: 29981422]
The genus Fusarium contains some of the most studied and important species of plant pathogens that economically affect world agriculture and horticulture. Fusarium spp. are ubiquitous fungi widely distributed in soil, plants as well as in different organic substrates and are also considered as opportunistic human pathogens. The identification of specific enzymes essential to the metabolism of these fungi is expected to provide molecular targets to control the diseases they induce to their hosts. Through applications of traditional techniques of sequence homology comparison by similarity search and Markov modeling, this report describes the characterization of enzymatic functionalities associated to protein targets that could be considered for the control of root rots induced by Fusarium oxysporum. From the analysis of 318 F. graminearum enzymes, we retrieved 30 enzymes that are specific of F. oxysporum compared to 15 species of host plants. By comparing these 30 specific enzymes of F. oxysporum with the genome of Arabidopsis thaliana, Brassica rapa, Glycine max, Jatropha curcas and Ricinus communis, we found 7 key specific enzymes whose inhibition is expected to affect significantly the development of the fungus and 5 specific enzymes that were considered here to be secondary because they are inserted in pathways with alternative routes.
Induction of Somatic Embryogenesis in Jatropha curcas.[Pubmed: 29981123]
Jatropha curcas has been a promising crop for biofuel production for the last decade. However, the lack of resistant materials to diseases and improved quality of the oil produced by the seeds has restricted the use of this promising crop. The genetic modifications in the fatty acid pathway, as well as the introduction of resistance to different diseases, would change the fate of Jatropha. To achieve these goals, we need to have a very efficient regeneration system. Here, we report a very useful protocol to induce somatic embryogenesis from leaves of Jatropha using cytokinin as the only growth regulator.


