Populus davidiana
Populus davidiana
1. The products in our compound library are selected from thousands of unique natural products; 2. It has the characteristics of diverse structure, diverse sources and wide coverage of activities; 3. Provide information on the activity of products from major journals, patents and research reports around the world, providing theoretical direction and research basis for further research and screening; 4. Free combination according to the type, source, target and disease of natural product; 5. The compound powder is placed in a covered tube and then discharged into a 10 x 10 cryostat; 6. Transport in ice pack or dry ice pack. Please store it at -20 °C as soon as possible after receiving the product, and use it as soon as possible after opening.
Natural products/compounds from Populus davidiana
- Cat.No. Product Name CAS Number COA
-
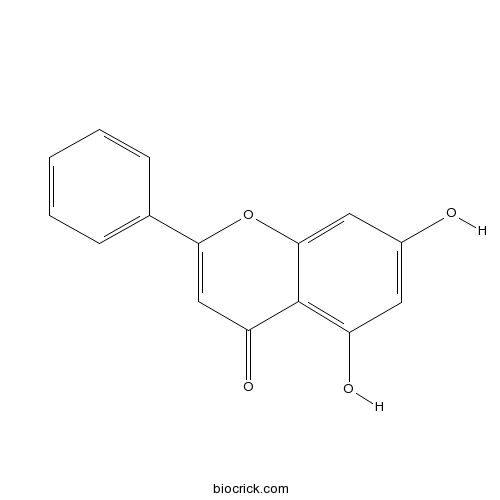
BCN5199
Sakuranetin2957-21-3
Instructions

-
BCN5488
Genkwanin437-64-9
Instructions

-
BCN5556
Pinocembrin480-39-7
Instructions

-
BCN5557
Chrysin480-40-0
Instructions
-
BCN5730
Galangin548-83-4
Instructions

Functional analysis of eliciting plant response protein Epl1-Tas from Trichoderma asperellum ACCC30536.[Pubmed: 29789617]
Eliciting plant response protein (Epl) is a small Trichoderma secreted protein that acts as an elicitor to induce plant defense responses against pathogens. In the present study, the differential expression, promoter analysis, and phylogenetic tree analysis of Epl1-Tas (GenBank JN966996) from T. asperellum ACCC30536 were performed. The results showed Epl1-Tas could play an important role in the interaction between T. asperellum ACCC30536 and woody plant or woody plant pathogen. Furthermore, the effect of the Escherichia coli recombinant protein rEpl1-e and the Pichia pastoris recombinant protein rEpl1-p on Populus davidiana × P. alba var. pyramidalis (PdPap) was studied. In PdPap seedlings, rEpl1-e or rEpl1-p induction altered the expression levels of 11 genes in the salicylic acid (SA, three genes), jasmonic acid (JA, four genes) and auxin (four genes) signal transduction pathways, and five kinds of enzymes activities The induction level of rEpl1-p was significantly higher than that of rEpl1-e, indicating that rEpl1-p could be used for further induction experiment. Under 3 mg/mL rEpl1-p induction, the mean height of the PdPap seedlings increased by 57.65% and the mean lesion area on the PdPap seedlings leaves challenged with Alternaria alternata decreased by 91.22% compared with those of the control. Thus, elicitor Epl1-Tas could induce the woody plant resistance to pathogen.
Rapid regeneration offsets losses from warming-induced tree mortality in an aspen-dominated broad-leaved forest in northern China.[Pubmed: 29624614]
Worldwide tree mortality as induced by climate change presents a challenge to forest managers. To successfully manage vulnerable forests requires the capacity of regeneration to compensate for losses from tree mortality. We observed rapid regeneration and the growth release of young trees after warming-induced mortality in a David aspen-dominated (Populus davidiana) broad-leaved forest in Inner Mongolia, China, as based on individual tree measurements taken in 2012 and 2015 from a 6-ha permanent plot. Warming and drought stress killed large trees 10-15 m tall with a total number of 2881 trees during 2011-2012, and also thinned the upper crowns. David aspen recruitment increased 2 times during 2012-2015 and resulted in a high transition probability of David aspen replacing the same or other species, whereas the recruitment of Mongolian oak (Quercus mongolica) was much lower: it decreased from 2012 to 2015, indicating that rapid regeneration represented a regrowth phase for David aspen, and not succession to Mongolian oak. Further, we found that the recruitment density increased with canopy openness, thus implying that warming-induced mortality enhanced regeneration. Our results suggest that David aspen has a high regrowth ability to offset individual losses from warming-induced mortality. This important insight has implications for managing this vulnerable forest in the semi-arid region of northern China.
Exploring genes involved in benzoic acid biosynthesis in the Populus davidiana transcriptome and their transcriptional activity upon methyl jasmonate treatment.[Pubmed: 29129016]
Benzoic acids (BAs) are important structural elements in a wide variety of essential compounds and natural products, and play crucial roles in plant fitness. BA is a precursor of diverse benzenoid compounds, including the hormone salicylic acid (SA) and the aglycone moiety of salicin, which is particularly important in the Salicaceae family. The biosynthetic pathways leading to BA formation in plants are largely unknown. Recently, the CoA-dependent β-oxidative BA biosynthesis pathway, which occurs in peroxisomes, has been characterized in petunia. The core of this pathway is cinnamic acid → cinnamoyl-CoA → 3-hydroxy-3-phenylpropanoyl-CoA → 3-oxo-3-phenylpropanoyl-CoA → benzoyl-CoA. Here, we used 454 pyrosequencing to analyze the transcriptome of Populus davidiana and isolate putative genes involved in BA biosynthesis. De novo assembly generated 57,322 unique sequences, including 15,217 contigs and 42,105 singletons. From the unique sequences, we selected six genes exhibiting high similarity to genes encoding L-phenylalanine ammonia lyase, cinnamate:CoA ligase, cinnamoyl-CoA hydratase-dehydrogenase, 3-ketoacyl-CoA thiolase, benzoyl-CoA:benzyl alcohol O-benzoyltransferase, and benzaldehyde dehydrogenase. Each of these enzymes might be involved in BA biosynthesis. Real-time PCR (qPCR) analysis revealed that these six genes were highly transcribed in the aerial organs of P. davidiana, particularly in leaves. Treating the leaves of in vitro cultured plants with methyl jasmonate (MeJA) strongly enhanced the mRNA accumulation of all 6 genes, and this treatment also clearly enhanced the accumulation of BA, SA, salicyl alcohol, benzyl alcohol, benzyl benzoate, and benzaldehyde but not salicin. Our study shows that P. davidiana may possess a CoA-dependent β-oxidative BA synthesis pathway. We also identified a relationship between the transcription of these genes and the accumulation of benzenoids, including BA and SA, which are highly responsive to the defense signaling molecule (MeJA).
Properties analysis of transcription factor gene TasMYB36 from Trichoderma asperellum CBS433.97 and its heterogeneous transfomation to improve antifungal ability of Populus.[Pubmed: 28993676]
The transcription of TasMYB36 in the biocontrol species T. asperellum was upregulated in four different pathogenic fermentation broths, suggesting that TasMYB36 plays an important role in the response to biotic stresses. Seventy-nine MYB transcription factors that were homologous to TasMYB36 from six sequenced Trichoderma genomes were analyzed. They were distributed in fourteen clades in the phylogenetic tree. The 79 MYBs contained 113 DNA binding domains, and their amino acid sequences were conserved and were different to those in plants. The promoters of 79 MYBs contained 1374 cis-regulators related to the stress response, such as GCR1 (17.5%) and GCN4 (15.5%). Subsequently, TasMYB36 was integrated into the genome of Populus davidiana × P. alba var. pyramidalis (PdPap poplar), and after co-culture of the transformants (PdPap-TasMYB36s) with Alternaria alternate, the transcription of genes in the jasmonic acid (JA) and salicylic acid (SA) hormone signaling pathways were upregulated; the POD, SOD and CAT activities were enhanced; and the reactive oxygen content was reduced in PdPap-TasMYB36s. The disease spots area on PdPap-TasMYB36s leaves infected by A. alternate were average 0.63% (PdPap-Con: 24.7%). In summary, TasMYB36 of T. asperellum CBS433.97 is an important defense response gene that upregulates other stress response genes and could improve resistance to biotic stresses.
Analysis of transcription factors among differentially expressed genes induced by drought stress in Populus davidiana.[Pubmed: 28667649]
Populus davidiana is native to the Korean Peninsula and is one of the most dominant and abundantly growing forest trees in eastern Asia. Compared to other Populus species such as P. trichocarpa, P. euphratica, and P. tremula, relatively little is known about P. davidiana. Here, we performed transcriptomic analysis of P. davidiana under drought stress induced by 10% polyethylene glycol. A total of 12,403 and 12,414 differentially expressed genes (DEGs) were successfully annotated with the P. trichocarpa reference genome after 6 and 12 h of treatment, respectively. Of these, a total of 404 genes (238 up-regulated and 166 down-regulated) after 6 h and 359 genes (187 up-regulated and 172 down-regulated) after 12 h of treatment were identified as transcription factors. Transcription factors known to be key genes for drought stress response, such as AP2-EREB, WRKY, C2H2, and NAC, were identified. This results suggesting that early induction of these genes affected initiation of transcriptional regulation in response to drought stress. Quantitative real-time PCR results of selected genes showed highly significant (R = 0.93) correlation with RNA-Seq data. Interestingly, the expression pattern of some transcription factors was P. davidiana specific. The sequence of P. davidiana ortholog of P. trichocarpa gene POPTR_0018s10230, which plays an important role in plant response to drought, was further analyzed as our RNA-Seq results showed highly significant changes in the expression of this gene following the stress treatment. Sequence of the gene was compared to P. trichocarpa gene sequence using cloning-based sequencing. Additionally, we generated a predicted 3D protein structure for the gene product. Results indicated that the amino acid sequence of P. davidiana-specific POPTR_0018s10230 is different at six different positions compared to P. trichocarpa, resulting in a significantly different structure of the protein. Identifying the transcription factors expressed in P. davidiana under drought stress will not only offer clues for understanding the underlying mechanisms involved in drought stress physiology but also serve as a basis for future molecular studies on this species.
[Research and Implementation of Wood Species Recognition System with Wood Near Infrared Spectral Reflection Features].[Pubmed: 30074341]
This paper proposes a novel wood species recognition scheme based on the spectral reflection features of wood surface, aiming to address the following three issues in terms of the noise filtering, feature selection and radian’s optimal design . First, noises occur in some bands of wood spectral reflection curve so that these noisy bands should be deleted. Second, the wood spectral band is 350~2 500 nm, which is a 2 150D vector with a spectral sampling interval of 1 nm. Therefore, both noise filtering and feature selection should be performed to wood spectral data. In this paper, to simultaneously and efficiently solve the two problems of feature selection and noise filtering, both a feature selection procedure and a noise filtering procedure are performed by solving the eigenvalues of dispersion matrix. This scheme is novel and produces a good outcome. Third, to make the spectral reflection curves picked up by the spectral instrument have the best pattern recognition information; an optimal design is performed for the indoor radian’s mounting height. The genetic algorithm is used to solve the optimal radian’s height so that the spectral reflection curves have the best classification information for wood species. Therefore, the optimal design scheme for the radian’s mounting height can improve the pattern classification accuracy of the wood species to some extents, which is novel with excellent executive feasibility. Many experiments made with our developed software system on the five ordinary wood species in northeast region of China (i.e., including Betula platyphylla, Populus davidiana, Pinus Sylvestris, Picea jezoensis, Larix gmelinii) are performed for approximately 105 times. It indicates that the overall recognition rate reaches to a good recognition accuracy of 95% for five wood species with an ideal recognition velocity. The selected feature wavelengths by using of our feature selection algorithm based on dispersion matrix are mainly in the near infrared band.


