Arachis hypogaea
Arachis hypogaea
1. The products in our compound library are selected from thousands of unique natural products; 2. It has the characteristics of diverse structure, diverse sources and wide coverage of activities; 3. Provide information on the activity of products from major journals, patents and research reports around the world, providing theoretical direction and research basis for further research and screening; 4. Free combination according to the type, source, target and disease of natural product; 5. The compound powder is placed in a covered tube and then discharged into a 10 x 10 cryostat; 6. Transport in ice pack or dry ice pack. Please store it at -20 °C as soon as possible after receiving the product, and use it as soon as possible after opening.
Natural products/compounds from Arachis hypogaea
- Cat.No. Product Name CAS Number COA
-
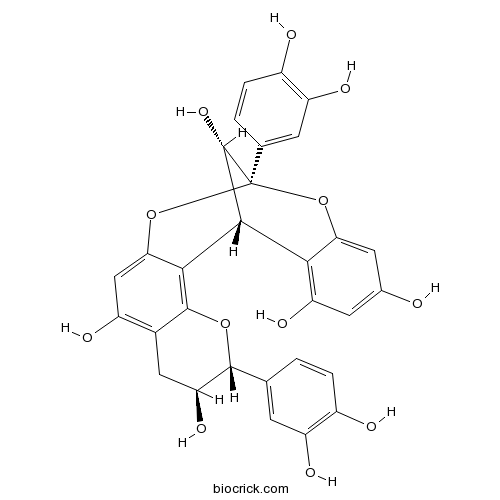
BCN6809
Procyanidin A1103883-03-0
Instructions
-
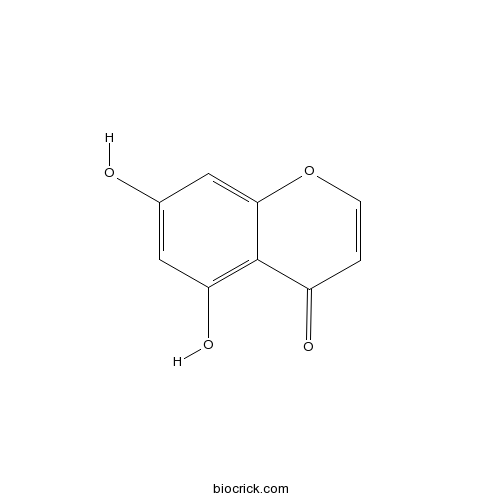
BCN4652
5,7-Dihydroxychromone31721-94-5
Instructions
-
BCN6805
Procyanidin A241743-41-3
Instructions

-
BCN5607
Resveratrol501-36-0
Instructions

-
BCN2356
Diosmetin520-34-3
Instructions

-
BCN1209
Eriodictyol552-58-9
Instructions

Transcriptomic analysis with the progress of symbiosis in 'crack-entry' legume Arachis hypogaea highlights its contrast with 'infection thread' adapted legumes.[Pubmed: 30109978]
In root-nodule symbiosis, rhizobial invasion and nodule organogenesis is host controlled. In most legumes, rhizobia enter through infection-threads and nodule primordium in the cortex is induced from a distance. But in dalbergoid legumes like Arachis hypogaea, rhizobia directly invade cortical cells through epidermal cracks to generate the primordia. Herein we report the transcriptional dynamics with the progress of symbiosis in A. hypogaea at 1dpi: invasion; 4dpi: nodule primordia; 8dpi: spread of infection in nodule-like structure; 12dpi: immature nodules containing rod-shaped rhizobia; and 21dpi: mature nodules with spherical symbiosomes. Expression of putative orthologue of symbiotic genes in 'crack-entry' legume A. hypogaea was compared with infection thread adapted model legumes. The contrasting features were (i) higher expression of receptors like LYR3, EPR3 as compared to canonical NFRs (ii) late induction of transcription factors like NIN, NSP2 and constitutive high expression of ERF1, EIN2, bHLH476 and (iii) induction of divergent pathogenesis responsive PR-1 genes. Additionally, symbiotic orthologues of SymCRK, ROP6, RR9, SEN1 and DNF2 were not detectable and microsynteny analysis indicated the absence of RPG homologue in diploid parental genomes of A. hypogaea. The implications are discussed and a molecular framework that guide 'crack-entry' symbiosis in A. hypogaea is proposed.
Genomic and Transcriptomic Analysis Identified Gene Clusters and Candidate Genes for Oil Content in Peanut (Arachis hypogaea L.).[Pubmed: 30100671]
None
Peanut.[Pubmed: 30000961]
Peanuts (Arachis hypogaea) contain carbohydrate, fat, and protein. Some of the proteins in peanuts are considered to be allergens that can lead to allergic reactions. Cooked, unripe peanuts are reportedly used in Africa and Asia as a galactogogue;[1] however, no scientifically valid clinical trials support this use. Galactogogues should never replace evaluation and counseling on modifiable factors that affect milk production.[2] Peanut protein allergens have been detected in breastmilk and some case reports and series have implicated maternal ingestion of peanuts during breastfeeding to peanut allergy in their breastfed infants.[3][4][5][6] Studies to determine whether maternal peanut ingestion during breastfeeding causes infant peanut allergy have come to conflicting conclusions. In the United States and United Kingdom, mothers were advised to avoid peanuts during pregnancy and breastfeeding in the late 1990s, but these recommendations have been withdrawn because of a lack of evidence of a reduction in peanut allergy.[7][8]


