Lygodium japonicum
Lygodium japonicum
1. The products in our compound library are selected from thousands of unique natural products; 2. It has the characteristics of diverse structure, diverse sources and wide coverage of activities; 3. Provide information on the activity of products from major journals, patents and research reports around the world, providing theoretical direction and research basis for further research and screening; 4. Free combination according to the type, source, target and disease of natural product; 5. The compound powder is placed in a covered tube and then discharged into a 10 x 10 cryostat; 6. Transport in ice pack or dry ice pack. Please store it at -20 °C as soon as possible after receiving the product, and use it as soon as possible after opening.
Natural products/compounds from Lygodium japonicum
- Cat.No. Product Name CAS Number COA
-
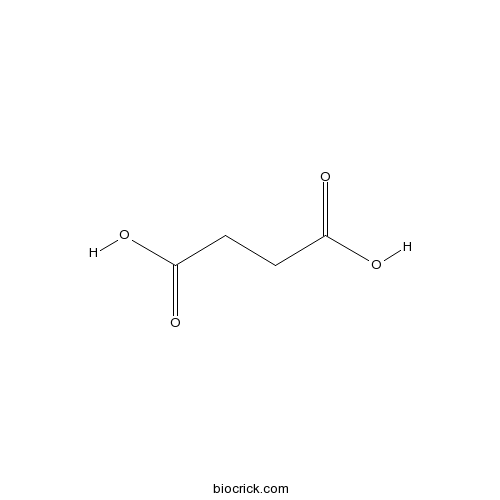
BCN5890
Succinic acid110-15-6
Instructions
-
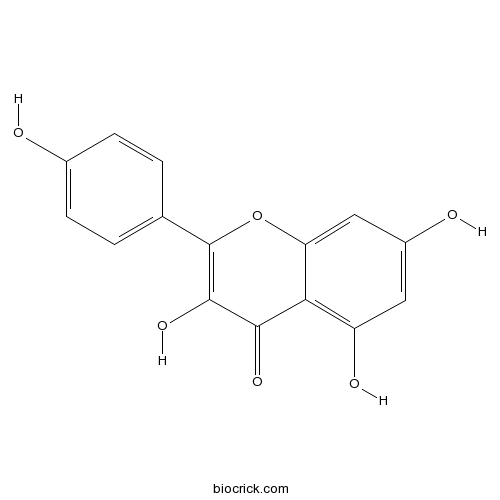
BCN5653
Kaempferol520-18-3
Instructions
-
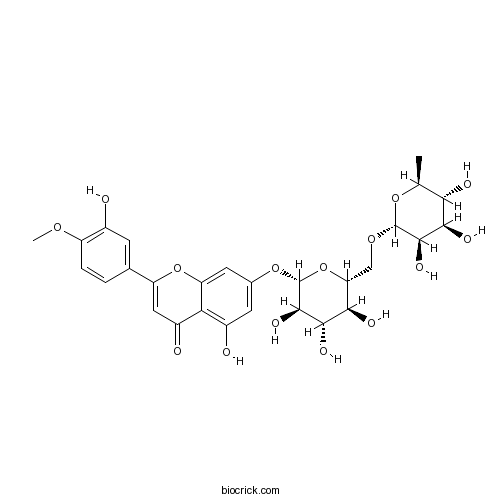
BCN4993
Diosimin520-27-4
Instructions
Anti‑inflammatory effects on murine macrophages of ethanol extracts of Lygodium japonicum spores via inhibition of NF‑κB and p38.[Pubmed: 29067444]
The spores of Lygodium japonicum (Thunb.) Sw. (L. japonicum) have been used in traditional Chinese medicine for the treatment of various inflammatory diseases. However, the molecular mechanisms underlying their anti‑inflammatory effects have yet to be elucidated. In the present study, we investigated the anti‑inflammatory effects of ethanol extracts of L. japonicum spores (ELJ) by measuring the production of inflammatory mediators, and explored the molecular mechanisms underlying the effects of ELJ in murine macrophages in vitro using immunoblotting analyses. At non‑cytotoxic concentrations of (50‑300 µg/ml), ELJ was revealed to significantly suppress the production of nitric oxide (NO) and tumor necrosis factor (TNF)‑α in lipopolysaccharide (LPS)‑stimulated murine RAW 264.7 macrophages; ELJ repressed the production of interleukin (IL)‑6 only at high concentrations (≥200 µg/ml). The ELJ‑mediated decrease in NO production was demonstrated to depend on the downregulation of inducible NO synthase mRNA and protein expression. Conversely, the mRNA and protein expression of cyclooxygenase‑2 were not affected by ELJ. In addition, ELJ was revealed to inhibit the mRNA expression of IL‑6, IL‑1β, and TNF‑α in LPS‑stimulated RAW 264.7 macrophages. The effects of ELJ on proinflammatory mediators may have been due to the stabilization of inhibitor of κBα and the inhibition of p38 mitogen‑activated protein kinase (MAPK). These results suggested that ELJ may suppress LPS‑induced inflammatory responses in murine macrophages in vitro, through the negative regulation of p38 MAPK and nuclear factor (NF)‑κB. Therefore, ELJ may have potential as a novel candidate for the development of therapeutic strategies aimed at alleviating inflammation.
Sequencing of small RNAs of the fern Pleopeltis minima (Polypodiaceae) offers insight into the evolution of the microrna repertoire in land plants.[Pubmed: 28494025]
MicroRNAs (miRNAs) are short, single stranded RNA molecules that regulate the stability and translation of messenger RNAs in diverse eukaryotic groups. Several miRNA genes are of ancient origin and have been maintained in the genomes of animal and plant taxa for hundreds of millions of years, playing key roles in development and physiology. In the last decade, genome and small RNA (sRNA) sequencing of several plant species have helped unveil the evolutionary history of land plants. Among these, the fern group (monilophytes) occupies a key phylogenetic position, as it represents the closest extant cousin taxon of seed plants, i.e. gymno- and angiosperms. However, in spite of their evolutionary, economic and ecological importance, no fern genome has been sequenced yet and few genomic resources are available for this group. Here, we sequenced the small RNA fraction of an epiphytic South American fern, Pleopeltis minima (Polypodiaceae), and compared it to plant miRNA databases, allowing for the identification of miRNA families that are shared by all land plants, shared by all vascular plants (tracheophytes) or shared by euphyllophytes (ferns and seed plants) only. Using the recently described transcriptome of another fern, Lygodium japonicum, we also estimated the degree of conservation of fern miRNA targets in relation to other plant groups. Our results pinpoint the origin of several miRNA families in the land plant evolutionary tree with more precision and are a resource for future genomic and functional studies of fern miRNAs.
Developmental morphology of the typical cordate gametophyte of a homosporous leptosporangiate fern, Lygodium japonicum (Lygodiaceae), focusing on the initial cell behavior of two distinct meristems.[Pubmed: 25667072]
Understanding the origin and early evolution of vascular plants requires thorough consideration of the gametophyte generation of ferns and lycophytes. Unfortunately, information about this generation is quite limited. To reveal the origin and evolution of varied gametophyte shapes, we used comparative morphological studies of meristem behavior of gametophytes of Lygodium japonicum, which exhibit the typical cordate shape.
De novo transcriptome assembly of a fern, Lygodium japonicum, and a web resource database, Ljtrans DB.[Pubmed: 25480117]
During plant evolution, ferns originally evolved as a major vascular plant with a distinctive life cycle in which the haploid and diploid generations are completely separated. However, the low level of genetic resources has limited studies of their physiological events, as well as hindering research on the evolutionary history of land plants. In this study, to identify a comprehensive catalog of transcripts and characterize their expression traits in the fern Lygodium japonicum, nine different RNA samples isolated from prothalli, trophophylls, rhizomes and sporophylls were sequenced using Roche 454 GS-FLX and Illumina HiSeq sequencers. The hybrid assembly of the high-quality 454 GS-FLX and Illumina HiSeq reads generated a set of 37,830 isoforms with an average length of 1,444 bp. Using four open reading frame (ORF) predictors, 38,142 representative ORFs were identified from a total of 37,830 transcript isoforms and 95 contigs, which were annotated by searching against several public databases. Furthermore, an orthoMCL analysis using the protein sequences of L. japonicum and five model plants revealed various sets of lineage-specific genes, including those detected among land plant lineages and those detected in only L. japonicum. We have also examined the expression patterns of all contigs/isoforms, along with the life cycle of L. japonicum, and identified the tissue-specific transcripts using statistical expression analyses. Finally, we developed a public web resource, the L. japonicum transcriptome database at http://bioinf.mind.meiji.ac.jp/kanikusa/, which provides important opportunities to accelerate molecular research in ferns.
The inhibitory effect of an ethanol extract of the spores of Lygodium japonicum on ethylene glycol-induced kidney calculi in rats.[Pubmed: 24972555]
We investigated the effect of an ethanol extract of Lygodii spora (LS) as a preventive and therapeutic agent for experimentally induced calcium oxalate nephrolithiasis with ethylene glycol (EG) in rats. Male Wistar rats were randomly divided into preventive (n = 18, for 28 days) and therapeutic (n = 24, for 42 days) groups. The preventive group was further subdivided into three groups of six rats each: preventive control, preventive lithiatic control (EG) and preventive lithiatic LS (EG + 400 mg/kg LS). Similarly, the therapeutic group was subdivided into four groups of six rats each: therapeutic control, therapeutic lithiatic control, therapeutic lithiatic untreated, and therapeutic lithiatic LS. Lithiasis was induced by adding 0.75% EG to the drinking water of all groups except the preventive and therapeutic control groups. Preventive and therapeutic subjects also received the LS ethanol extract in drinking water at a dose of 400 mg/kg, since day 0 or day 28, respectively. At the end of the each experimental period, various biochemical parameters were measured in urine and kidney homogenates. The kidneys were subjected to histopathological analysis. The results revealed that treatment with the LS preventive protocol significantly decreased the levels of urinary calcium, oxalate and uric acid, and increased the levels of urinary citrate as compared to those in the EG control. No significant changes in the urinary parameters except oxalate and citrate levels were observed in the rats in the therapeutic protocol. In both preventive and therapeutic protocols, the extract significantly decreased kidney peroxides, renal calcium, oxalate content, and the number of kidney oxalate deposits as compared to those in the EG group. We conclude that LS is useful as a preventive and therapeutic agent against the formation of oxalate kidney stones.
Chloroplast genome evolution in early diverged leptosporangiate ferns.[Pubmed: 24823358]
In this study, the chloroplast (cp) genome sequences from three early diverged leptosporangiate ferns were completed and analyzed in order to understand the evolution of the genome of the fern lineages. The complete cp genome sequence of Osmunda cinnamomea (Osmundales) was 142,812 base pairs (bp). The cp genome structure was similar to that of eusporangiate ferns. The gene/intron losses that frequently occurred in the cp genome of leptosporangiate ferns were not found in the cp genome of O. cinnamomea. In addition, putative RNA editing sites in the cp genome were rare in O. cinnamomea, even though the sites were frequently predicted to be present in leptosporangiate ferns. The complete cp genome sequence of Diplopterygium glaucum (Gleicheniales) was 151,007 bp and has a 9.7 kb inversion between the trnL-CAA and trnVGCA genes when compared to O. cinnamomea. Several repeated sequences were detected around the inversion break points. The complete cp genome sequence of Lygodium japonicum (Schizaeales) was 157,142 bp and a deletion of the rpoC1 intron was detected. This intron loss was shared by all of the studied species of the genus Lygodium. The GC contents and the effective numbers of codons (ENCs) in ferns varied significantly when compared to seed plants. The ENC values of the early diverged leptosporangiate ferns showed intermediate levels between eusporangiate and core leptosporangiate ferns. However, our phylogenetic tree based on all of the cp gene sequences clearly indicated that the cp genome similarity between O. cinnamomea (Osmundales) and eusporangiate ferns are symplesiomorphies, rather than synapomorphies. Therefore, our data is in agreement with the view that Osmundales is a distinct early diverged lineage in the leptosporangiate ferns.
Plastome sequences of Lygodium japonicum and Marsilea crenata reveal the genome organization transformation from basal ferns to core leptosporangiates.[Pubmed: 23821521]
Previous studies have shown that core leptosporangiates, the most species-rich group of extant ferns (monilophytes), have a distinct plastid genome (plastome) organization pattern from basal fern lineages. However, the details of genome structure transformation from ancestral ferns to core leptosporangiates remain unclear because of limited plastome data available. Here, we have determined the complete chloroplast genome sequences of Lygodium japonicum (Lygodiaceae), a member of schizaeoid ferns (Schizaeales), and Marsilea crenata (Marsileaceae), a representative of heterosporous ferns (Salviniales). The two species represent the sister and the basal lineages of core leptosporangiates, respectively, for which the plastome sequences are currently unavailable. Comparative genomic analysis of all sequenced fern plastomes reveals that the gene order of L. japonicum plastome occupies an intermediate position between that of basal ferns and core leptosporangiates. The two exons of the fern ndhB gene have a unique pattern of intragenic copy number variances. Specifically, the substitution rate heterogeneity between the two exons is congruent with their copy number changes, confirming the constraint role that inverted repeats may play on the substitution rate of chloroplast gene sequences.
Unusual 9,19 : 24,32-dicyclotetracyclic triterpenoids from Lygodium japonicum.[Pubmed: 23154839]
Lygodipenoids A (1) and B (2), two novel C33 tetracyclic triterpenoids with a new 9,19 : 24,32-dicyclopropane skeleton, were isolated from the whole grass of Lygodium japonicum. Their structures were elucidated by spectroscopic and chemical means. Compounds 1 and 2 were tested in transfected cultured human embryonic kidney 293 HEK293 cells for an agonist assay, and compound 1 was identified as a partial agonist for liver X receptor α.
A new phenylpropanoid glucoside from the aerial parts of Lygodium japonicum.[Pubmed: 22251217]
A new compound, 4-O-caffeoyl-D-glucopyranose (1), and a new natural product, 3-O-caffeoyl-D-glucopyranose (2), together with six known compounds (3-8) were isolated from the aerial parts of Lygodium japonicum. Their structures were elucidated on the basis of extensive spectroscopic data analyses. The oxygen radical absorbance capacity assay was applied to evaluate their antioxidative capacities in vitro, which revealed that 1-8 showed strong antioxidative properties.
Nicotinamide metabolism in ferns: formation of nicotinic acid glucoside.[Pubmed: 21251843]
The metabolic fate of [carbonyl-(14)C]nicotinamide was investigated in 9 fern species, Psilotum nudum, Angiopteris evecta, Lygodium japonicum, Acrostichum aureum, Asplenium antiquum, Diplazium subsinuatum, Thelypteris acuminate, Blechnum orientale and Crytomium fortune. All fern species produce a large quantity of nicotinic acid glucoside from [(14)C]nicotinamide, but trigonelline formation is very low. Increases in the release of (14)CO(2) with incubation time was accompanied by decreases in [carboxyl-(14)C]nicotinic acid glucoside. There was slight stimulation of nicotinic acid glucoside formation by 250 mM NaCl in mature leaves of the mangrove fern, Acrostichum aureum, but it is unlikely that this compound acts as a compatible solute. Nicotinamide and nicotinic acid salvage for pyridine nucleotide synthesis was detected in all fern species, although this activity was always less than nicotinic acid glucoside synthesis. Predominant formation of nicotinic acid glucoside is characteristic of nicotinic acid metabolism in ferns. This reaction appears to act as a detoxication mechanism, removing excess nicotinic acid.


