N,N',N''-TriacetylchitotrioseCAS# 38864-21-0 |
Quality Control & MSDS
3D structure
Package In Stock
Number of papers citing our products

| Cas No. | 38864-21-0 | SDF | Download SDF |
| PubChem ID | 134693119.0 | Appearance | Powder |
| Formula | C24H41N3O16 | M.Wt | 627.6 |
| Type of Compound | Oligoses | Storage | Desiccate at -20°C |
| Solubility | Soluble in Chloroform,Dichloromethane,Ethyl Acetate,DMSO,Acetone,etc. | ||
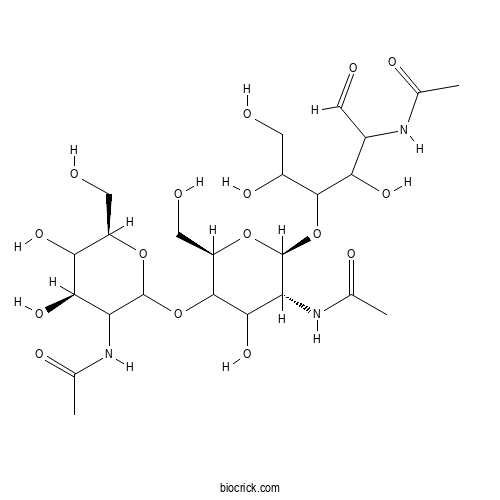
| Chemical Name | N-[(4R,6R)-2-[(2R,5R,6S)-5-acetamido-6-(5-acetamido-1,2,4-trihydroxy-6-oxohexan-3-yl)oxy-4-hydroxy-2-(hydroxymethyl)oxan-3-yl]oxy-4,5-dihydroxy-6-(hydroxymethyl)oxan-3-yl]acetamide | ||
| SMILES | CC(=O)NC1C(C(C(OC1OC2C(OC(C(C2O)NC(=O)C)OC(C(CO)O)C(C(C=O)NC(=O)C)O)CO)CO)O)O | ||
| Standard InChIKey | LRDDKCYRFNJZBX-YLFBAVNJSA-N | ||
| Standard InChI | InChI=1S/C24H41N3O16/c1-8(32)25-11(4-28)17(36)21(12(35)5-29)42-24-16(27-10(3)34)20(39)22(14(7-31)41-24)43-23-15(26-9(2)33)19(38)18(37)13(6-30)40-23/h4,11-24,29-31,35-39H,5-7H2,1-3H3,(H,25,32)(H,26,33)(H,27,34)/t11?,12?,13-,14-,15?,16-,17?,18?,19-,20?,21?,22?,23?,24+/m1/s1 | ||
| General tips | For obtaining a higher solubility , please warm the tube at 37 ℃ and shake it in the ultrasonic bath for a while.Stock solution can be stored below -20℃ for several months. We recommend that you prepare and use the solution on the same day. However, if the test schedule requires, the stock solutions can be prepared in advance, and the stock solution must be sealed and stored below -20℃. In general, the stock solution can be kept for several months. Before use, we recommend that you leave the vial at room temperature for at least an hour before opening it. |
||
| About Packaging | 1. The packaging of the product may be reversed during transportation, cause the high purity compounds to adhere to the neck or cap of the vial.Take the vail out of its packaging and shake gently until the compounds fall to the bottom of the vial. 2. For liquid products, please centrifuge at 500xg to gather the liquid to the bottom of the vial. 3. Try to avoid loss or contamination during the experiment. |
||
| Shipping Condition | Packaging according to customer requirements(5mg, 10mg, 20mg and more). Ship via FedEx, DHL, UPS, EMS or other couriers with RT, or blue ice upon request. | ||

N,N',N''-Triacetylchitotriose Dilution Calculator

N,N',N''-Triacetylchitotriose Molarity Calculator
| 1 mg | 5 mg | 10 mg | 20 mg | 25 mg | |
| 1 mM | 1.5934 mL | 7.9669 mL | 15.9337 mL | 31.8674 mL | 39.8343 mL |
| 5 mM | 0.3187 mL | 1.5934 mL | 3.1867 mL | 6.3735 mL | 7.9669 mL |
| 10 mM | 0.1593 mL | 0.7967 mL | 1.5934 mL | 3.1867 mL | 3.9834 mL |
| 50 mM | 0.0319 mL | 0.1593 mL | 0.3187 mL | 0.6373 mL | 0.7967 mL |
| 100 mM | 0.0159 mL | 0.0797 mL | 0.1593 mL | 0.3187 mL | 0.3983 mL |
| * Note: If you are in the process of experiment, it's necessary to make the dilution ratios of the samples. The dilution data above is only for reference. Normally, it's can get a better solubility within lower of Concentrations. | |||||

Calcutta University

University of Minnesota

University of Maryland School of Medicine

University of Illinois at Chicago

The Ohio State University

University of Zurich

Harvard University

Colorado State University

Auburn University

Yale University

Worcester Polytechnic Institute

Washington State University

Stanford University

University of Leipzig

Universidade da Beira Interior

The Institute of Cancer Research

Heidelberg University

University of Amsterdam

University of Auckland

TsingHua University

The University of Michigan

Miami University

DRURY University

Jilin University

Fudan University

Wuhan University

Sun Yat-sen University

Universite de Paris

Deemed University

Auckland University

The University of Tokyo

Korea University
- N,N',N'',N'''-Tetraacetylchitotetraose
Catalog No.:BCX1414
CAS No.:2706-65-2
- N,N',N'',N''',N''''-Pentaacetyl chitopentaose
Catalog No.:BCX1413
CAS No.:36467-68-2
- N,N',N'',N''',N'''',N'''''-Hexaacetyl chitohexaose
Catalog No.:BCX1412
CAS No.:38854-46-5
- Chitobiose Dihydrochloride
Catalog No.:BCX1411
CAS No.:115350-24-8
- Chitotetraose Tetrahydrochloride
Catalog No.:BCX1410
CAS No.:117399-50-5
- Chitopentaose Pentahydrochloride
Catalog No.:BCX1409
CAS No.:117467-64-8
- Chitohexaose Hexahydrochloride
Catalog No.:BCX1408
CAS No.:127171-88-4
- 5-Alpha-Hydroxy Laxogenin
Catalog No.:BCX1407
CAS No.:56786-63-1
- 1β-Methoxydiversifolin 3-O-methyl ether
Catalog No.:BCX1406
CAS No.:194474-71-0
- Emetine hydrobromide
Catalog No.:BCX1405
CAS No.:52714-87-1
- 2'-Deoxyguanosine monohydrate
Catalog No.:BCX1404
CAS No.:312693-72-4
- Gylongiposide I
Catalog No.:BCX1403
CAS No.:206876-12-2
- N,N'-Diacetylchitobiose
Catalog No.:BCX1416
CAS No.:35061-50-8
- Chitopentaose Pentadecaacetate
Catalog No.:BCX1417
CAS No.:117399-52-7
- Chitotetraose tetradecaacetate
Catalog No.:BCX1418
CAS No.:53942-46-4
- Chitotriose Undecaacetate
Catalog No.:BCX1419
CAS No.:53942-45-3
- Chitobiose Octaacetate
Catalog No.:BCX1420
CAS No.:41670-99-9
- Chitotriose Trihydrochloride
Catalog No.:BCX1421
CAS No.:117436-78-9
- Chitooctaose Octahydrochloride
Catalog No.:BCX1422
CAS No.:127171-90-8
- Chitoheptaose Heptahydrochloride
Catalog No.:BCX1423
CAS No.:127171-89-5
- L-octaguluronic acid octasodium salt
Catalog No.:BCX1424
CAS No.:862694-88-0
- L-heptaguluronic acid heptasodium salt
Catalog No.:BCX1425
CAS No.:862694-87-9
- L-hexaguluronic acid hexasodium salt
Catalog No.:BCX1426
CAS No.:183668-74-8
- L-pentaguluronic acid pentasodium salt
Catalog No.:BCX1427
CAS No.:183668-72-6
Hallmarks of Comparative Transcriptome between Rhizomorphs and Hyphae of Armillaria sp. 541 Participating in Fungal Symbiosis with Emphasis on LysM Domains.[Pubmed:37630474]
Microorganisms. 2023 Jul 27;11(8):1914.
Armillaria sp. 541, a genus of root-infecting fungi, forms a symbiosis with traditional Chinese medicine Gastrodia elata (Orchid) and Polyporus umbellatus via extensive networks of durable rhizomorphs. It is not clear the hallmarks of comparative transcriptome between the rhizomorphs and hyphae of Armillaria sp. 541. In the present study, transcriptomic analysis of Armillaria sp. 541 identified 475 differentially expressed genes (DEGs) between Armillaria rhizomorphs (AR) and hyphae (AH). Of them, 285 genes were upregulated and 190 were downregulated. Bioinformatics analyses and tests demonstrated DEGs involved in oxidoreductase activity and peptidoglycan binding were significantly enriched in this process when rhizomorph formed from hyphae. We accordingly obtained 14 gene-encoding proteins containing the LysM domain, and further consensus pattern and phylogenetic analysis indicated that their amino acid sequences were conserved and their biological functions may be peptidoglycan binding for recognition between the fungus and host. Among these genes, one, named Armillaria LysM domain recognition gene (aLDRG), was expressed significantly when rhizomorphs were differentiated from hyphae. It was located in the cortical cells of the rhizomorph by in situ hybridization. Furthermore, biolayer interferometry (BLI) assay demonstrated that aLDRG can bind specifically to chitin oligosaccharide of the fungal cell wall, including N,N',N''-Triacetylchitotriose (CO3) and N,N',N'',N''',N''''-Pentaacetylchitopentaose (CO5). Therefore, we deduced that Armillaria sp. 541 expressed higher levels of LysM protein aLDRG for better binding of oligosaccharide after rhizomorphs were generated. This study provides functional genes for further studies on the interaction between Armillaria sp. 541 and its host.
Chitin, Chitin Oligosaccharide, and Chitin Disaccharide Metabolism of Escherichia coli Revisited: Reassignment of the Roles of ChiA, ChbR, ChbF, and ChbG.[Pubmed:33794535]
Microb Physiol. 2021;31(2):178-194.
Escherichia coli is unable to grow on polymeric and oligomeric chitin, but grows on chitin disaccharide (GlcNAc-GlcNAc; N,N'-diacetylchitobiose) and chitin trisaccharide (GlcNAc-GlcNAc-GlcNAc; N,N',N''-triacetylchitotriose) via expression of the chb operon (chbBCARFG). The phosphotransferase system (PTS) transporter ChbBCA facilitates transport of both saccharides across the inner membrane and their concomitant phosphorylation at the non-reducing end, intracellularly yielding GlcNAc 6-phosphate-GlcNAc (GlcNAc6P-GlcNAc) and GlcNAc6P-GlcNAc-GlcNAc, respectively. We revisited the intracellular catabolism of the PTS products, thereby correcting the reported functions of the 6-phospho-glycosidase ChbF, the monodeacetylase ChbG, and the transcriptional regulator ChbR. Intracellular accumulation of glucosamine 6P-GlcNAc (GlcN6P-GlcNAc) and GlcN6P-GlcNAc-GlcNAc in a chbF mutant unraveled a role for ChbG as a monodeacetylase that removes the N-acetyl group at the non-reducing end. Consequently, GlcN6P- but not GlcNAc6P-containing saccharides likely function as coactivators of ChbR. Furthermore, ChbF removed the GlcN6P from the non-reducing terminus of the former saccharides, thereby degrading the inducers of the chb operon and facilitating growth on the saccharides. Consequently, ChbF was unable to hydrolyze GlcNAc6P-residues from the non-reducing end, contrary to previous assumptions but in agreement with structural modeling data and with the unusual catalytic mechanism of the family 4 of glycosidases, to which ChbF belongs. We also refuted the assumption that ChiA is a bifunctional endochitinase/lysozyme ChiA, and show that it is unable to degrade peptidoglycans but acts as a bona fide chitinase in vitro and in vivo, enabling growth of E. coli on chitin oligosaccharides when ectopically expressed. Overall, this study revises our understanding of the chitin, chitin oligosaccharide, and chitin disaccharide metabolism of E. coli.
Detecting Protein-Ligand Interaction from Integrated Transient Induced Molecular Electronic Signal (i-TIMES).[Pubmed:32045225]
Anal Chem. 2020 Mar 3;92(5):3852-3859.
Quantitative information about protein-ligand interactions is central to drug discovery. To obtain the quintessential reaction dissociation constant, ideally measurements of reactions should be performed without perturbations by molecular labeling or immobilization. The technique of transient induced molecular electrical signal (TIMES) has provided a promising technique to meet such requirements, and its performance in a microfluidic environment further offers the potential for high throughput and reduced consumption of reagents. In this work, we further the development by using integrated TIMES signal (i-TIMES) to greatly enhance the accuracy and reproducibility of the measurement. While the transient response may be of interest, the integrated signal directly measures the total amount of surface charge density resulted from molecules near the surface of electrode. The signals enable quantitative characterization of protein-ligand interactions. We have demonstrated the feasibility of i-TIMES technique using different biomolecules including lysozyme, N,N',N''-triacetylchitotriose (TriNAG), aptamer, p-aminobenzamidine (pABA), bovine pancreatic ribonuclease A (RNaseA), and uridine-3'-phosphate (3'UMP). The results show i-TIMES is a simple and accurate technique that can bring tremendous value to drug discovery and research of intermolecular interactions.
Computational modeling and functional characterization of a GgChi: A class III chitinase from corms of Gladiolus grandiflorus.[Pubmed:30527201]
Kaohsiung J Med Sci. 2018 Dec;34(12):673-683.
The present study describes the predicted model and functional characterization of an endochitinase (30 kDa) from corms of Gladiolus grandiflorus. ESI-QTOF-MS generated peptide showed 96% sequence homology with family 18, Class III acidic endochitinase of Gladiolus gandavensis. Purified G. grandiflorus chitinase (GgChi) hydrolyzed 4-methylumbelliferyl beta-d-N,N',N''-triacetylchitotriose substrate showing specific endochitinase activity. Since no structural details of GgChi were available in the Protein Data Bank (PDB), a homology model was predicted using the coordinate information of Crocus vernus chitinase (PDB ID: 3SIM). Ramachandran plot indicated 84.5% in most favored region, 14.8% in additional and 0.6% in generously allowed region while no residue in disallowed region. The predicted structure indicated a highly conserved (beta/alpha)(8) (TIM barrel) structure similar to the family 18, class III chitinases. The GgChi also showed sequence and structural homologies with other active chitinases. The GgChi (50 mug/disc) showed no antibacterial activity, but did provide mild growth inhibition of phytopathogenic fungus Fusarium oxysporum at a concentration of 500 mug/well Similarly, insect toxicity bioassays of GgChi (50 mug) against nymphs of Bemisia tabaci showed 14% reduction in adult emergence and 14% increase in mortality rate in comparison to control values. The GgChi (1.5 mg) protein showed significant reduction in a population of flour beetle (Tribolium castaneum) after 35 days, but lower reactivity against rice weevil (Sitophilus oryzae). The results of this study provide detai.led insight on functional characterization of a family 18 class III acidic plant endochitinase.
Probing specific ligand-protein interactions by native-denatured exchange mass spectrometry.[Pubmed:30253837]
Anal Chim Acta. 2018 Dec 7;1036:58-65.
Probing ligand-target protein interactions provides essential information for deep understanding of biochemical machinery and design of drug screening assays. Native electrospray ionization-mass spectrometry (ESI-MS) is promising for direct analysis of ligand-protein complexes. However, it lacks the ability to distinguish between specific and non-specific ligand-protein interactions, and to further recognize the specifically bound proteins as drug target candidates, which remains as a major challenge in the field of drug developments by far. Herein we report a native-denatured exchange (NDX) mass spectrometry (MS) acquisition approach using a liquid sample-desorption electrospray ionization (LS-DESI) setup, and demonstrate its capability in enabling a change from native detection of noncovalent ligand-protein complexes to denatured analysis using three model ligand-protein complexes including myoglobin, CDP-ribonuclease and N,N',N''-triacetylchitotriose (NAG3)-lysozyme. Notably, we found the NDX-MS approach can readily discriminate specific ligand-protein interactions from nonspecific ones, as revealed by their distinct dynamic profiles of K(d) as a function of the DESI spraying flow rate. Consequently, this NDX-MS approach holds promise for future applications to discovering specific protein targets for ligands of interest, and to screening compounds with high specificity to drug targets and thus eliminates off-target effects.
Enzymatic hydrolysis of ionic liquid-extracted chitin.[Pubmed:30143125]
Carbohydr Polym. 2018 Nov 1;199:228-235.
Chitin, one of Nature's most abundant biopolymers, can be obtained by either traditional chemical pulping or by extraction using the ionic liquid (IL) 1-ethyl-3-methylimidazolium acetate. The IL extraction and coagulation process provides access to a unique chitin, with an open hydrated gel-like structure. Here, enzymatic hydrolysis of this chitin hydrogel, dried shrimp shell, chitin extracted from shrimp shells using IL and then dried, and commercial chitin was carried out using chitinase from Streptomyces griseus. The enzymatic hydrolysis of shrimp shells resulted only in the monomer N-acetylglucosamine, while much higher amounts of the dimer (N,N'-diacetylchitobiose) compared to the monomer were detected when using all forms of 'pure' chitin. Interestingly, small amounts of the trimer (N,N',N''-triacetylchitotriose) were also detected when the IL-chitin hydrogel was used as substrate. Altogether, our findings indicate that the product distribution and yield are highly dependent on the substrate selected for the reaction and its hydrated state.
Cloning and characterization of a chitinase from Thermobifida fusca reveals Tfu_0580 as a thermostable and acidic endochitinase.[Pubmed:30094208]
Biotechnol Rep (Amst). 2018 Jul 14;19:e00274.
Being capable of hydrolyzing chitin, chitinases have various applications such as production of N-acetylchitooligosaccharides (COSs) and N-acetylglucosamine (GlcNAc), degrading chitin as a consolidated bioprocessing, and bio-control of fungal phytopathogens. Here, a putative chitinase in Thermobifida fusca, Tfu_0580, is characterized. Tfu_0580 was purified by homogeneity with a molecular weight of 44.9 kDa by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) analysis. Tfu_0580 displayed a clear activity against colloidal chitin, which is comparable to a commercial Streptomyces griseus chitinase. Enzyme activities against p-nitrophenyl beta-D-N,N',N''-triacetylchitotriose (p-NP-(GlcNAc)(3)), N,N'-diacetyl-beta-D-chitobioside (p-NP-(GlcNAc)(2)) and p-nitrophenyl N-acetyl-beta-D-glucosaminide (p-NP-(GlcNAc)) showed that Tfu_0580 exhibited highest activity against p-NP-(GlcNAc)(3). Further optimization of the enzyme activity conditions showed: 1) an optimum catalytic activity at pH 6.0 and 30 degrees C; 2) activity over broad pH (4.8-7.5) and temperature (20-55 degrees C); 3) stimulation of activity by the metallic ions Ca(2+) and Mn(2+).
Quantification of Protein-Ligand Interactions by Laser Electrospray Mass Spectrometry.[Pubmed:29654537]
J Am Soc Mass Spectrom. 2018 Jul;29(7):1484-1492.
Laser electrospray mass spectrometry (LEMS) measurement of the dissociation constant (K(d)) for hen egg white lysozyme (HEWL) and N,N',N''-triacetylchitotriose (NAG(3)) revealed an apparent K(d) value of 313.2 +/- 25.9 muM for the ligand titration method. Similar measurements for N,N',N'',N'''-tetraacetylchitotetraose (NAG(4)) revealed an apparent K(d) of 249.3 +/- 13.6 muM. An electrospray ionization mass spectrometry (ESI-MS) experiment determined a K(d) value of 9.8 +/- 0.6 muM. In a second LEMS approach, a calibrated measurement was used to determine a K(d) value of 6.8 +/- 1.5 muM for NAG(3). The capture efficiency of LEMS was measured to be 3.6 +/- 1.8% and is defined as the fraction of LEMS sample detected after merging with the ESI plume. When the dilution is factored into the ligand titration measurement, the adjusted K(d) value was 11.3 muM for NAG(3) and 9.0 muM for NAG(4). The calibration method for measuring K(d) developed in this study can be applied to solutions containing unknown analyte concentrations. Graphical Abstract.


