Trifolium pratense
Trifolium pratense
1. The products in our compound library are selected from thousands of unique natural products; 2. It has the characteristics of diverse structure, diverse sources and wide coverage of activities; 3. Provide information on the activity of products from major journals, patents and research reports around the world, providing theoretical direction and research basis for further research and screening; 4. Free combination according to the type, source, target and disease of natural product; 5. The compound powder is placed in a covered tube and then discharged into a 10 x 10 cryostat; 6. Transport in ice pack or dry ice pack. Please store it at -20 °C as soon as possible after receiving the product, and use it as soon as possible after opening.

Natural products/compounds from Trifolium pratense
- Cat.No. Product Name CAS Number COA
-
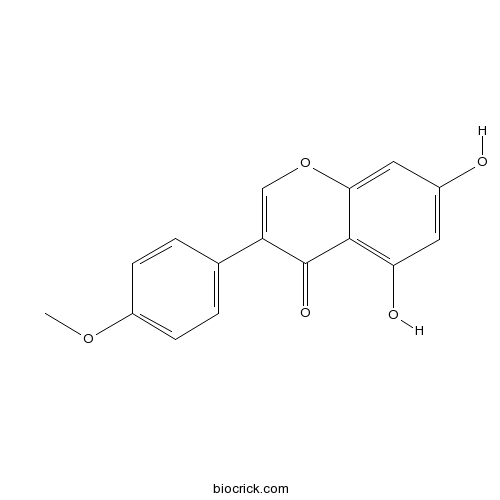
BCN5499
Genistein446-72-0
Instructions

-
BCN1061
Formononetin485-72-3
Instructions

-
BCN5926
Ononin486-62-4
Instructions

-
BCN5590
Daidzein486-66-8
Instructions

-
BCN1224
Biochanin A491-80-5
Instructions
-
BCN2396
Genistin529-59-9
Instructions

-
BCN5891
Daidzin552-66-9
Instructions

-
BCN4237
Trifolirhizin6807-83-6
Instructions

-
BCC4109
Salicylic acid69-72-7
Instructions

-
BCN4327
Ursolic acid77-52-1
Instructions

Comparison of intracellular location and stimulus reaction times of forisomes in sieve tubes of four legume species.[Pubmed: 30111246]
Forisomes in legumes are responsible for fast sieve-element occlusion in response to injury to the vascular system. This prevents uncontrolled leakage of phloem sap and protects against invasion of pathogens. Here we compared forisomes of four different legumes (Pisum sativum, Vicia faba, Trifolium pratense and Medicago sativa) by their location (basal, central, apical) in the sieve element and reactivity to a distant heat stimulus. In each species, the majority of forisomes was located basally. Yet, we found differences in intracellular location: forisomes are distributed more evenly in the sieve elements of Pisum. After burning, basally located forisomes of the four species reacted with dispersion, followed by a spontaneous recondensation with similar reaction times. The results suggest universal forisome behaviour in fabacean species.
Genome-wide atlas of alternative polyadenylation in the forage legume red clover.[Pubmed: 30054540]
Studies on prevalence and significance of alternative polyadenylation (APA) in plants have been so far limited mostly to the model plants. Here, a genome-wide analysis of APA was carried out in different tissue types in the non-model forage legume red clover (Trifolium pratense L). A profile of poly(A) sites in different tissue types was generated using so-called 'poly(A)-tag sequencing' (PATseq) approach. Our analysis revealed tissue-wise dynamics of usage of poly(A) sites located at different genomic locations. We also identified poly(A) sites and underlying genes displaying APA in different tissues. Functional categories enriched in groups of genes manifesting APA between tissue types were determined. Analysis of spatial expression of genes encoding different poly(A) factors showed significant differential expression of genes encoding orthologs of FIP1(V) and PCFS4, suggesting that these two factors may play a role in regulating spatial APA in red clover. Our analysis also revealed a high degree of conservation in diverse plant species of APA events in mRNAs encoding two key polyadenylation factors, CPSF30 and FIP1(V). Together with our previously reported study of spatial gene expression in red clover, this study will provide a comprehensive account of transcriptome dynamics in this non-model forage legume.


