Negsehisandrin GCAS# 1023744-69-5 |
Quality Control & MSDS
3D structure
Package In Stock
Number of papers citing our products

| Cas No. | 1023744-69-5 | SDF | Download SDF |
| PubChem ID | 23583761 | Appearance | Powder |
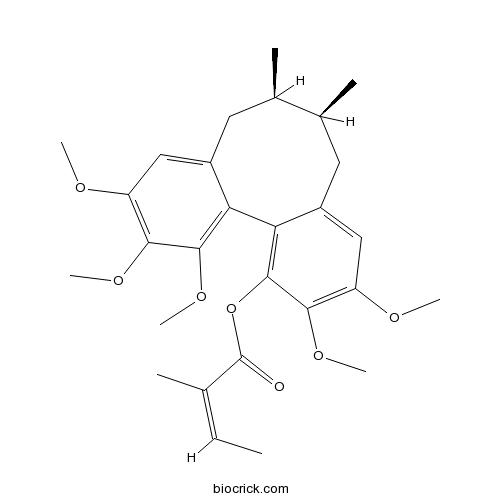
| Formula | C28H36O7 | M.Wt | 484.6 |
| Type of Compound | Lignans | Storage | Desiccate at -20°C |
| Solubility | Soluble in Chloroform,Dichloromethane,Ethyl Acetate,DMSO,Acetone,etc. | ||
| SMILES | CC=C(C)C(=O)OC1=C2C(=CC(=C1OC)OC)CC(C(CC3=CC(=C(C(=C32)OC)OC)OC)C)C | ||
| Standard InChIKey | DSAHZJYWMDAZSA-KNUIFBHBSA-N | ||
| Standard InChI | InChI=1S/C28H36O7/c1-10-15(2)28(29)35-27-23-19(14-21(31-6)25(27)33-8)12-17(4)16(3)11-18-13-20(30-5)24(32-7)26(34-9)22(18)23/h10,13-14,16-17H,11-12H2,1-9H3/b15-10-/t16-,17+/m1/s1 | ||
| General tips | For obtaining a higher solubility , please warm the tube at 37 ℃ and shake it in the ultrasonic bath for a while.Stock solution can be stored below -20℃ for several months. We recommend that you prepare and use the solution on the same day. However, if the test schedule requires, the stock solutions can be prepared in advance, and the stock solution must be sealed and stored below -20℃. In general, the stock solution can be kept for several months. Before use, we recommend that you leave the vial at room temperature for at least an hour before opening it. |
||
| About Packaging | 1. The packaging of the product may be reversed during transportation, cause the high purity compounds to adhere to the neck or cap of the vial.Take the vail out of its packaging and shake gently until the compounds fall to the bottom of the vial. 2. For liquid products, please centrifuge at 500xg to gather the liquid to the bottom of the vial. 3. Try to avoid loss or contamination during the experiment. |
||
| Shipping Condition | Packaging according to customer requirements(5mg, 10mg, 20mg and more). Ship via FedEx, DHL, UPS, EMS or other couriers with RT, or blue ice upon request. | ||

Negsehisandrin G Dilution Calculator

Negsehisandrin G Molarity Calculator
| 1 mg | 5 mg | 10 mg | 20 mg | 25 mg | |
| 1 mM | 2.0636 mL | 10.3178 mL | 20.6356 mL | 41.2712 mL | 51.5889 mL |
| 5 mM | 0.4127 mL | 2.0636 mL | 4.1271 mL | 8.2542 mL | 10.3178 mL |
| 10 mM | 0.2064 mL | 1.0318 mL | 2.0636 mL | 4.1271 mL | 5.1589 mL |
| 50 mM | 0.0413 mL | 0.2064 mL | 0.4127 mL | 0.8254 mL | 1.0318 mL |
| 100 mM | 0.0206 mL | 0.1032 mL | 0.2064 mL | 0.4127 mL | 0.5159 mL |
| * Note: If you are in the process of experiment, it's necessary to make the dilution ratios of the samples. The dilution data above is only for reference. Normally, it's can get a better solubility within lower of Concentrations. | |||||

Calcutta University

University of Minnesota

University of Maryland School of Medicine

University of Illinois at Chicago

The Ohio State University

University of Zurich

Harvard University

Colorado State University

Auburn University

Yale University

Worcester Polytechnic Institute

Washington State University

Stanford University

University of Leipzig

Universidade da Beira Interior

The Institute of Cancer Research

Heidelberg University

University of Amsterdam

University of Auckland

TsingHua University

The University of Michigan

Miami University

DRURY University

Jilin University

Fudan University

Wuhan University

Sun Yat-sen University

Universite de Paris

Deemed University

Auckland University

The University of Tokyo

Korea University
- Naringin
Catalog No.:BCN6312
CAS No.:10236-47-2
- Cadensin D
Catalog No.:BCN7260
CAS No.:102349-35-9
- (R)-Eriodictyol-8-C-beta-D-glucopyranoside
Catalog No.:BCN8028
CAS No.:1023271-51-3
- Epoxyparvinolide
Catalog No.:BCN5838
CAS No.:102227-61-2
- SGX-523
Catalog No.:BCC1055
CAS No.:1022150-57-7
- Btk inhibitor 1 R enantiomer
Catalog No.:BCC5125
CAS No.:1022150-12-4
- ARN2966
Catalog No.:BCC8074
CAS No.:102212-26-0
- Tyrosine kinase inhibitor
Catalog No.:BCC2020
CAS No.:1021950-26-4
- Boc-4-oxo-Pro-OMe
Catalog No.:BCC3436
CAS No.:102195-80-2
- 3-O-Methyltirotundin
Catalog No.:BCN5837
CAS No.:1021945-29-8
- ZM 39923 HCl
Catalog No.:BCC2203
CAS No.:1021868-92-7
- AGN 192403 hydrochloride
Catalog No.:BCC6924
CAS No.:1021868-90-5
- Glyburide
Catalog No.:BCC4784
CAS No.:10238-21-8
- AF-DX 116
Catalog No.:BCC6939
CAS No.:102394-31-0
- Jasplakinolide
Catalog No.:BCC7485
CAS No.:102396-24-7
- Fmoc-Phg-OH
Catalog No.:BCC3312
CAS No.:102410-65-1
- RN 1747
Catalog No.:BCC7769
CAS No.:1024448-59-6
- Kushenol N
Catalog No.:BCN2984
CAS No.:102490-65-3
- GK921
Catalog No.:BCC8057
CAS No.:1025015-40-0
- Periglaucine A
Catalog No.:BCN5839
CAS No.:1025023-04-4
- Periglaucine B
Catalog No.:BCN7053
CAS No.:1025023-05-5
- SGI-1776 free base
Catalog No.:BCC2232
CAS No.:1025065-69-3
- (-)-Huperzine A
Catalog No.:BCN1057
CAS No.:102518-79-6
- 2,3,23-Trihydroxy-12-oleanen-28-oic acid
Catalog No.:BCN1638
CAS No.:102519-34-6
Expression, Functional Characterization, and Solid-State NMR Investigation of the G Protein-Coupled GHS Receptor in Bilayer Membranes.[Pubmed:28387359]
Sci Rep. 2017 Apr 7;7:46128.
The expression, functional reconstitution and first NMR characterization of the human growth hormone secretagogue (GHS) receptor reconstituted into either DMPC or POPC membranes is described. The receptor was expressed in E. coli. refolded, and reconstituted into bilayer membranes. The molecule was characterized by (15)N and (13)C solid-state NMR spectroscopy in the absence and in the presence of its natural agonist ghrelin or an inverse agonist. Static (15)N NMR spectra of the uniformly labeled receptor are indicative of axially symmetric rotational diffusion of the G protein-coupled receptor in the membrane. In addition, about 25% of the (15)N sites undergo large amplitude motions giving rise to very narrow spectral components. For an initial quantitative assessment of the receptor mobility, (1)H-(13)C dipolar coupling values, which are scaled by molecular motions, were determined quantitatively. From these values, average order parameters, reporting the motional amplitudes of the individual receptor segments can be derived. Average backbone order parameters were determined with values between 0.56 and 0.69, corresponding to average motional amplitudes of 40-50 degrees of these segments. Differences between the receptor dynamics in DMPC or POPC membranes were within experimental error. Furthermore, agonist or inverse agonist binding only insignificantly influenced the average molecular dynamics of the receptor.
G-protein coupled receptor 15 mediates angiogenesis and cytoprotective function of thrombomodulin.[Pubmed:28386128]
Sci Rep. 2017 Apr 6;7(1):692.
Thrombomodulin (TM) stimulates angiogenesis and protects vascular endothelial cells (ECs) via its fifth epidermal growth factor-like region (TME5); however, the cell surface receptor that mediates the pro-survival signaling activated by TM has remained unknown. We applied pull-down assay followed by MALDI-TOF MS and western blot analysis, and identified G-protein coupled receptor 15 (GPR15) as a binding partner of TME5. TME5 rescued growth inhibition and apoptosis caused by calcineurin inhibitor FK506 in vascular ECs isolated from wild type (WT) C57BL/6 mice. On the other hand, TME5 failed to protect ECs isolated from GPR15 knockout (GPR15 KO) mice from FK506-caused vascular injury. TME5 induced activation of extracellular signal-regulated kinase (ERK) and increased level of anti-apoptotic proteins in a GPR15 dependent manner. In addition, in vivo Matrigel plug angiogenesis assay found that TME5 stimulated angiogenesis in mice. TME5 promoted endothelial migration in vitro. Furthermore, TME5 increased production of NO in association with activated endothelial NO synthase (eNOS) in ECs. All these pro-angiogenesis functions of TME5 were abolished by knockout of GPR15. Our findings suggest that GPR15 plays an important role in mediating cytoprotective function as well as angiogenesis of TM.
Cancer-associated noncoding mutations affect RNA G-quadruplex-mediated regulation of gene expression.[Pubmed:28386116]
Sci Rep. 2017 Apr 6;7(1):708.
Cancer is a multifactorial disease driven by a combination of genetic and environmental factors. Many cancer driver mutations have been characterised in protein-coding regions of the genome. However, mutations in noncoding regions associated with cancer have been less investigated. G-quadruplex (G4) nucleic acids are four-stranded secondary structures formed in guanine-rich sequences and prevalent in the regulatory regions. In this study, we used published whole cancer genome sequence data to find mutations in cancer patients that overlap potential RNA G4-forming sequences in 5' UTRs. Using RNAfold, we assessed the effect of these mutations on the thermodynamic stability of predicted RNA G4s in the context of full-length 5' UTRs. Of the 217 identified mutations, we found that 33 are predicted to destabilise and 21 predicted to stabilise potential RNA G4s. We experimentally validated the effect of destabilising mutations in the 5' UTRs of BCL2 and CXCL14 and one stabilising mutation in the 5' UTR of TAOK2. These mutations resulted in an increase or a decrease in translation of these mRNAs, respectively. These findings suggest that mutations that modulate the G4 stability in the noncoding regions could act as cancer driver mutations, which present an opportunity for early cancer diagnosis using individual sequencing information.


