GinnoCAS# 2606-50-0 |
Quality Control & MSDS
3D structure
Package In Stock
Number of papers citing our products

| Cas No. | 2606-50-0 | SDF | Download SDF |
| PubChem ID | 16057860.0 | Appearance | Powder |
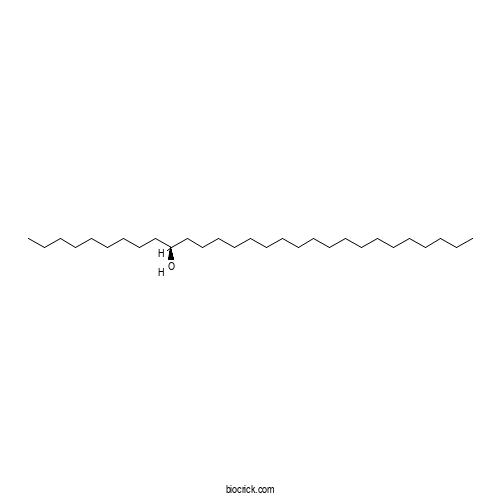
| Formula | C29H60O | M.Wt | 424.8 |
| Type of Compound | N/A | Storage | Desiccate at -20°C |
| Solubility | Soluble in Chloroform,Dichloromethane,Ethyl Acetate,DMSO,Acetone,etc. | ||
| Chemical Name | (10S)-nonacosan-10-ol | ||
| SMILES | CCCCCCCCCCCCCCCCCCCC(CCCCCCCCC)O | ||
| Standard InChIKey | CPGCVOVWHCWVTP-LJAQVGFWSA-N | ||
| Standard InChI | InChI=1S/C29H60O/c1-3-5-7-9-11-12-13-14-15-16-17-18-19-20-22-24-26-28-29(30)27-25-23-21-10-8-6-4-2/h29-30H,3-28H2,1-2H3/t29-/m0/s1 | ||
| General tips | For obtaining a higher solubility , please warm the tube at 37 ℃ and shake it in the ultrasonic bath for a while.Stock solution can be stored below -20℃ for several months. We recommend that you prepare and use the solution on the same day. However, if the test schedule requires, the stock solutions can be prepared in advance, and the stock solution must be sealed and stored below -20℃. In general, the stock solution can be kept for several months. Before use, we recommend that you leave the vial at room temperature for at least an hour before opening it. |
||
| About Packaging | 1. The packaging of the product may be reversed during transportation, cause the high purity compounds to adhere to the neck or cap of the vial.Take the vail out of its packaging and shake gently until the compounds fall to the bottom of the vial. 2. For liquid products, please centrifuge at 500xg to gather the liquid to the bottom of the vial. 3. Try to avoid loss or contamination during the experiment. |
||
| Shipping Condition | Packaging according to customer requirements(5mg, 10mg, 20mg and more). Ship via FedEx, DHL, UPS, EMS or other couriers with RT, or blue ice upon request. | ||

Ginno Dilution Calculator

Ginno Molarity Calculator
| 1 mg | 5 mg | 10 mg | 20 mg | 25 mg | |
| 1 mM | 2.354 mL | 11.7702 mL | 23.5405 mL | 47.081 mL | 58.8512 mL |
| 5 mM | 0.4708 mL | 2.354 mL | 4.7081 mL | 9.4162 mL | 11.7702 mL |
| 10 mM | 0.2354 mL | 1.177 mL | 2.354 mL | 4.7081 mL | 5.8851 mL |
| 50 mM | 0.0471 mL | 0.2354 mL | 0.4708 mL | 0.9416 mL | 1.177 mL |
| 100 mM | 0.0235 mL | 0.1177 mL | 0.2354 mL | 0.4708 mL | 0.5885 mL |
| * Note: If you are in the process of experiment, it's necessary to make the dilution ratios of the samples. The dilution data above is only for reference. Normally, it's can get a better solubility within lower of Concentrations. | |||||

Calcutta University

University of Minnesota

University of Maryland School of Medicine

University of Illinois at Chicago

The Ohio State University

University of Zurich

Harvard University

Colorado State University

Auburn University

Yale University

Worcester Polytechnic Institute

Washington State University

Stanford University

University of Leipzig

Universidade da Beira Interior

The Institute of Cancer Research

Heidelberg University

University of Amsterdam

University of Auckland

TsingHua University

The University of Michigan

Miami University

DRURY University

Jilin University

Fudan University

Wuhan University

Sun Yat-sen University

Universite de Paris

Deemed University

Auckland University

The University of Tokyo

Korea University
- LINOLELAIDICACIDMETHYLESTER
Catalog No.:BCX0936
CAS No.:2566-97-4
- Coumarin 343
Catalog No.:BCX0935
CAS No.:55804-65-4
- Hydroxypinacolone Retinoate
Catalog No.:BCX0934
CAS No.:893412-73-2
- 2-Methoxybenzoicacid
Catalog No.:BCX0933
CAS No.:529-75-9
- Nicotinamidemononucleotide
Catalog No.:BCX0932
CAS No.:1094-61-7
- Cepharanoline
Catalog No.:BCX0931
CAS No.:27686-34-6
- PhysalinF
Catalog No.:BCX0930
CAS No.:57517-46-1
- 3-(3-hydroxylphenyl)propanol
Catalog No.:BCX0929
CAS No.:621-54-5
- (+)-Pinoresinolmonomethylether4-O-β-D-glucoside
Catalog No.:BCX0928
CAS No.:74957-57-6
- Hydroxyisogermafurenolide
Catalog No.:BCX0927
CAS No.:20267-91-8
- 14-Deoxy-11-oxoandrographolide
Catalog No.:BCX0926
CAS No.:42895-57-8
- MogrosideIIA
Catalog No.:BCX0925
CAS No.:1613527-65-3
- β-Elemene
Catalog No.:BCX0938
CAS No.:515-13-9
- PhlorigidosideC
Catalog No.:BCX0939
CAS No.:276691-32-8
- (S)-(-)-Norcoclaurinehydrobromide
Catalog No.:BCX0940
CAS No.:105990-27-0
- 14-hydroxylatedbrassinosteroid
Catalog No.:BCX0941
CAS No.:457603-63-3
- Harmalinehydrochloridedihydrate
Catalog No.:BCX0942
CAS No.:6027-98-1
- Lycobetaineacetate
Catalog No.:BCX0943
CAS No.:61221-41-8
- CompoundK
Catalog No.:BCX0944
CAS No.:160729-91-9
- 11-hydroxy-1-isomangostin
Catalog No.:BCX0945
CAS No.:164365-71-3
- GarcixanthonesB
Catalog No.:BCX0946
CAS No.:2522597-99-3
- MangostanaxanthoneIV
Catalog No.:BCX0947
CAS No.:2182593-73-1
- Carpesiolin
Catalog No.:BCX0948
CAS No.:63568-73-0
- Aromaticin
Catalog No.:BCX0949
CAS No.:5945-42-6
Exploration and analysis of R-loop mapping data with RLBase.[Pubmed:36039757]
Nucleic Acids Res. 2023 Jan 6;51(D1):D1129-D1137.
R-loops are three-stranded nucleic acid structures formed from the hybridization of RNA and DNA. In 2012, Ginno et al. introduced the first R-loop mapping method. Since that time, dozens of R-loop mapping studies have been conducted, yielding hundreds of publicly available datasets. Current R-loop databases provide only limited access to these data. Moreover, no web tools for analyzing user-supplied R-loop datasets have yet been described. In our recent work, we reprocessed 810 R-loop mapping samples, building the largest R-loop data resource to date. We also defined R-loop consensus regions and developed a framework for R-loop data analysis. Now, we introduce RLBase, a user-friendly database that provides the capability to (i) explore hundreds of public R-loop mapping datasets, (ii) explore R-loop consensus regions, (iii) analyze user-supplied data and (iv) download standardized and reprocessed datasets. RLBase is directly accessible via the following URL: https://gccri.bishop-lab.uthscsa.edu/shiny/rlbase/.
Detection and Characterization of R Loop Structures.[Pubmed:28349431]
Methods Mol Biol. 2017;1543:231-242.
R loops are special three stranded nucleic acid structures that comprise a nascent RNA hybridized with the DNA template strand, leaving a non-template DNA single-stranded. More specifically, R loops form in vivo as G-rich RNA transcripts invade the DNA duplex and anneal to the template strand to generate an RNA:DNA hybrid, leaving the non-template, G-rich DNA strand in a largely single-stranded conformation (Aguilera and Garcia-Muse, Mol Cell 46:115-124, 2012).DNA-RNA hybrids are a natural occurrence within eukaryotic cells, with levels of these hybrids increasing at sites with high transcriptional activity, such as during transcription initiation, repression, and elongation. RNA-DNA hybrids influence genomic instability, and growing evidence points to an important role for R loops in active gene expression regulation (Ginno et al., Mol Cell 45, 814-825, 2012; Sun et al., Science 340: 619-621, 2013; Bhatia et al., Nature 511, 362-365, 2014). Analysis of the occurrence of such structures is therefore of increasing relevance and herein we describe methods for the in vivo and in vitro identification and characterization of R loops in mammalian systems.R loops (DNA:RNA hybrids and the associated single-stranded DNA) have been traditionally associated with threats to genome integrity, making some regions of the genome more prone to DNA-damaging and mutagenic agents. Initially considered to be rare byproducts of transcription, over the last decade accumulating evidence has pointed to a new view in which R loops form more frequently than previously thought. The R loop field has become an increasingly expanded area of research, placing these structures as a major threat to genome stability but also as potential regulators of gene expression. Special interest has arisen as they have also been linked to a variety of diseases, including neurological disorders and cancer, positioning them as potential therapeutic targets [5].
RNA-FISH to Study Regulatory RNA at the Site of Transcription.[Pubmed:28349430]
Methods Mol Biol. 2017;1543:221-229.
The increasing role of all types of regulatory RNAs in the orchestration of cellular programs has enhanced the development of a variety of techniques that allow its precise detection, quantification, and functional scrutiny. Recent advances in imaging and fluoresecent in situ hybridization (FISH) methods have enabled the utilization of user-friendly protocols that provide highly sensitive and accurate detection of ribonucleic acid molecules at both the single cell and subcellular levels. We herein describe the approach originally developed by Stellaris((R)), in which the target RNA molecule is fluoresecently labeled with multiple tiled complementary probes each carrying a fluorophore, thus improving sensitivity and reducing the chance of false positives. We have applied this method to the detection of nascent RNAs that partake of special regulatory structures called R loops. Their growing role in active gene expression regulation (Aguilera and Garcia-Muse, Mol Cell 46:115-124, 2012; Ginno et al., Mol Cell 45:814-825, 2012; Sun et al., Science 340:619-621, 2013; Bhatia et al., Nature 511:362-365, 2014) imposes the use of a combination of in vivo and in vitro techniques for the detailed analysis of the transcripts involved. Therefore, their study is a good example to illustrate how RNA FISH, combined with transcriptional arrest and/or cell synchronization, permits localization and temporal characterization of potentially regulatory RNA sequences.
R loops: lassoing DNA methylation at CpGi.[Pubmed:22464440]
Mol Cell. 2012 Mar 30;45(6):708-9.
In this issue of Molecular Cell, Ginno et al. (2012) describe unusual sequence features at promoter CpG islands that can lead to formation of persistent RNA-DNA hybrids (R loops), which are proposed to prevent genomic DNA methylation.


