L189inhibitor of human DNA ligases I, III and IV CAS# 64232-83-3 |
- Dexpramipexole dihydrochloride
Catalog No.:BCC1528
CAS No.:104632-27-1
- Dexpramipexole
Catalog No.:BCC1527
CAS No.:104632-28-2
- Cariprazine hydrochloride
Catalog No.:BCC1454
CAS No.:1083076-69-0
- Cariprazine
Catalog No.:BCC1453
CAS No.:839712-12-8
Quality Control & MSDS
3D structure
Package In Stock
Number of papers citing our products

| Cas No. | 64232-83-3 | SDF | Download SDF |
| PubChem ID | 824710 | Appearance | Powder |
| Formula | C11H10N4OS | M.Wt | 246.29 |
| Type of Compound | N/A | Storage | Desiccate at -20°C |
| Solubility | DMSO : ≥ 33 mg/mL (133.99 mM) *"≥" means soluble, but saturation unknown. | ||
| Chemical Name | 6-amino-5-(benzylideneamino)-2-sulfanylidene-1H-pyrimidin-4-one | ||
| SMILES | C1=CC=C(C=C1)C=NC2=C(NC(=S)NC2=O)N | ||
| Standard InChIKey | SAKOXVNKDMWWLF-UHFFFAOYSA-N | ||
| Standard InChI | InChI=1S/C11H10N4OS/c12-9-8(10(16)15-11(17)14-9)13-6-7-4-2-1-3-5-7/h1-6H,(H4,12,14,15,16,17) | ||
| General tips | For obtaining a higher solubility , please warm the tube at 37 ℃ and shake it in the ultrasonic bath for a while.Stock solution can be stored below -20℃ for several months. We recommend that you prepare and use the solution on the same day. However, if the test schedule requires, the stock solutions can be prepared in advance, and the stock solution must be sealed and stored below -20℃. In general, the stock solution can be kept for several months. Before use, we recommend that you leave the vial at room temperature for at least an hour before opening it. |
||
| About Packaging | 1. The packaging of the product may be reversed during transportation, cause the high purity compounds to adhere to the neck or cap of the vial.Take the vail out of its packaging and shake gently until the compounds fall to the bottom of the vial. 2. For liquid products, please centrifuge at 500xg to gather the liquid to the bottom of the vial. 3. Try to avoid loss or contamination during the experiment. |
||
| Shipping Condition | Packaging according to customer requirements(5mg, 10mg, 20mg and more). Ship via FedEx, DHL, UPS, EMS or other couriers with RT, or blue ice upon request. | ||
| Description | DNA ligase I, III and IV inhibitor (IC50 values are 5, 9 and 5 μM respectively) that blocks DNA binding (Ki = 5 μM for DNA ligase I). Preferentially inhibits step 2 of the ligation reaction, and inhibits base excision repair (BER) and non-homologous end joining (NHEJ). Specifically sensitizes cancer cells to DNA damage and increases the cytotoxicity of DNA-damaging agents. |

L189 Dilution Calculator

L189 Molarity Calculator
| 1 mg | 5 mg | 10 mg | 20 mg | 25 mg | |
| 1 mM | 4.0603 mL | 20.3013 mL | 40.6025 mL | 81.2051 mL | 101.5064 mL |
| 5 mM | 0.8121 mL | 4.0603 mL | 8.1205 mL | 16.241 mL | 20.3013 mL |
| 10 mM | 0.406 mL | 2.0301 mL | 4.0603 mL | 8.1205 mL | 10.1506 mL |
| 50 mM | 0.0812 mL | 0.406 mL | 0.8121 mL | 1.6241 mL | 2.0301 mL |
| 100 mM | 0.0406 mL | 0.203 mL | 0.406 mL | 0.8121 mL | 1.0151 mL |
| * Note: If you are in the process of experiment, it's necessary to make the dilution ratios of the samples. The dilution data above is only for reference. Normally, it's can get a better solubility within lower of Concentrations. | |||||

Calcutta University

University of Minnesota

University of Maryland School of Medicine

University of Illinois at Chicago

The Ohio State University

University of Zurich

Harvard University

Colorado State University

Auburn University

Yale University

Worcester Polytechnic Institute

Washington State University

Stanford University

University of Leipzig

Universidade da Beira Interior

The Institute of Cancer Research

Heidelberg University

University of Amsterdam

University of Auckland

TsingHua University

The University of Michigan

Miami University

DRURY University

Jilin University

Fudan University

Wuhan University

Sun Yat-sen University

Universite de Paris

Deemed University

Auckland University

The University of Tokyo

Korea University
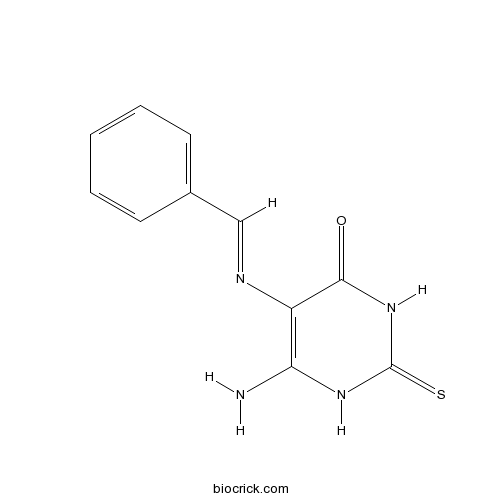
The chemical structure of L189 had been shown [1]. Three distinct chemical steps are involved in the formation of a new phosphodiester bond in DNA: (1) enzyme adenylylation, (2) adenylyl transfer to DNA, and (3) nick sealing [2]. L189 preferentially inhibits the second step [1]. L189 showed no inhibition even at a concentration of 100 μM to T4 DNA ligase. IC50 values for L189 to inhibit human DNA ligases I, III and IV are 5 ± 2 µM, 9 ± 2 µM and 5 ± 2 µM, respectively [1].
DNA ligase I catalyzes the ligation of single-strand breaks to complete DNA replication and repair. DNA ligase III is required for mitochondrial DNA replication and repair. DNA ligase IV is specialized for repair of nuclear double-strand breaks and is required for nonhomologous end joining and V (D) J recombination [2].
In a concentration-dependent manner, L189 reduced the viability and/or proliferation of a normal breast epithelial cell line MCF10A and the cancer cell lines HeLa, MCF7 and HCT116 established from cervical, breast and colon cancers, respectively. L189 also increase the rate of killed cells in cancer cell lines, especially HCT116 colon cancer cell lines and other cancer cell lines by ionizing radiation. But L189 did not increase the rate of killed cells in the normal cell line [1].
In vivo data and clinical trials are not yet available [3].
References:
[1]. Xi Chen, Shijun Zhong, Xiao Zhu, et al. Rational Design of Human DNA Ligase Inhibitors that Target Cellular DNA Replication and Repair. Cancer Res., 2008, 68(9): 3169-3177.
[2]. Mark R. Taylor, John A. Conrad, Daniel Wahl, et al. Kinetic Mechanism of Human DNA Ligase I Reveals Magnesium-dependent Changes in the Rate-limiting Step That Compromise Ligation Efficiency. Journal of Biological Chemistry, 2011, 286(26): 23054-23062.
[3]. Christian Jekimovs, Emma Bolderson, Amila Suraweera, et al. Chemotherapeutic compounds targeting the DNA double-strand break repair pathways: the good, the bad, and the promising. Frontiers in Oncology, 2014, 4: Article 86.
- Atracurium oxalate
Catalog No.:BCC8837
CAS No.:64228-78-0
- Thalirugidine
Catalog No.:BCN7706
CAS No.:64215-95-8
- CGP 12177 hydrochloride
Catalog No.:BCC6949
CAS No.:64208-32-8
- Antiarol
Catalog No.:BCN4185
CAS No.:642-71-7
- L-Quebrachitol
Catalog No.:BCN2727
CAS No.:642-38-6
- Alstonine
Catalog No.:BCN4606
CAS No.:642-18-2
- Akuammigine
Catalog No.:BCN4607
CAS No.:642-17-1
- Alloimperatorin
Catalog No.:BCC8116
CAS No.:642-05-7
- Barbinervic acid
Catalog No.:BCN4061
CAS No.:64199-78-6
- Senaetnine
Catalog No.:BCN2127
CAS No.:64191-69-1
- Zanthobungeanine
Catalog No.:BCN6685
CAS No.:64190-94-9
- Z-Hyp-Ome
Catalog No.:BCC3258
CAS No.:64187-48-0
- Taraxasterol acetate
Catalog No.:BCN4184
CAS No.:6426-43-3
- Glutinol acetate
Catalog No.:BCN6675
CAS No.:6426-44-4
- Boc-N-Me-Tyr(Bzl)-OH
Catalog No.:BCC3356
CAS No.:64263-81-6
- Kielcorin
Catalog No.:BCN7637
CAS No.:64280-48-4
- Tetrahydropapaverine HCl
Catalog No.:BCC5321
CAS No.:6429-04-5
- Acetagastrodin
Catalog No.:BCN8155
CAS No.:64291-41-4
- Galactopinitol A
Catalog No.:BCC8926
CAS No.:64290-91-1
- Dracorhodin
Catalog No.:BCC9226
CAS No.:643-56-1
- Malvidin chloride
Catalog No.:BCN3017
CAS No.:643-84-5
- N6-Benzoyl-5'-O-(4,4'-dimethoxytrityl)-2'-deoxyadenosine
Catalog No.:BCC9074
CAS No.:64325-78-6
- Alloalantolactone
Catalog No.:BCN8091
CAS No.:64340-41-6
- Precocene II
Catalog No.:BCN4605
CAS No.:644-06-4
Structure-activity relationships among DNA ligase inhibitors: Characterization of a selective uncompetitive DNA ligase I inhibitor.[Pubmed:29078112]
DNA Repair (Amst). 2017 Dec;60:29-39.
In human cells, there are three genes that encode DNA ligase polypeptides with distinct but overlapping functions. Previously small molecule inhibitors of human DNA ligases were identified using a structure-based approach. Three of these inhibitors, L82, a DNA ligase I (LigI)-selective inhibitor, and L67, an inhibitor of LigI and DNA ligases III (LigIII), and L189, an inhibitor of all three human DNA ligases, have related structures that are composed of two 6-member aromatic rings separated by different linkers. Here we have performed a structure-activity analysis to identify determinants of activity and selectivity. The majority of the LigI-selective inhibitors had a pyridazine ring whereas the LigI/III- and LigIII-selective inhibitors did not. In addition, the aromatic rings in LigI-selective inhibitors had either arylhydrazone or acylhydrazone, but not vinyl linkers. Among the LigI-selective inhibitors, L82-G17 exhibited increased activity against and selectivity for LigI compared with L82. Notably. L82-G17 is an uncompetitive inhibitor of the third step of the ligation reaction, phosphodiester bond formation. Cells expressing LigI were more sensitive to L82-G17 than isogenic LIG1 null cells. Furthermore, cells lacking nuclear LigIIIalpha, which can substitute for LigI in DNA replication, were also more sensitive to L82-G17 than isogenic parental cells. Together, our results demonstrate that L82-G17 is a LigI-selective inhibitor with utility as a probe of the catalytic activity and cellular functions of LigI and provide a framework for the future design of DNA ligase inhibitors.
Positive selection in bone morphogenetic protein 15 targets a natural mutation associated with primary ovarian insufficiency in human.[Pubmed:24147118]
PLoS One. 2013 Oct 16;8(10):e78199.
Bone Morphogenetic Protein 15 (BMP15) is a TGFbeta-like oocyte-derived growth factor involved in ovarian folliculogenesis as a critical regulator of many granulosa cell processes. Alterations of the BMP15 gene have been found associated with different ovarian phenotypic effects depending on the species, from sterility to increased prolificacy in sheep, slight subfertility in mouse or associated with primary ovarian insufficiency (POI) in women. To investigate the evolving role of BMP15, a phylogenetic analysis of this particular TGFbeta family member was performed. A maximum likelihood phylogenetic tree of several TGFbeta/BMP family members expressed by the ovary showed that BMP15 has a very strong divergence and a rapid evolution compared to others. Moreover, among 24 mammalian species, we detected signals of positive selection in the hominidae clade corresponding to F146, L189 and Y235 residues in human BMP15. The biological importance of these residues was tested functionally after site directed-mutagenesis in a COV434 cells luciferase assay. By replacing the positively selected amino acid either by alanine or the most represented residue in other studied species, only L189A, Y235A and Y235C mutants showed a significant increase of BMP15 signaling when compared to wild type. Additionally, the Y235C mutant was more potent than wild type in inhibiting progesterone secretion of ovine granulosa cells in primary culture. Interestingly, the Y235C mutation was previously identified in association with POI in women. In conclusion, this study evidences that the BMP15 gene has evolved faster than other members of the TGFss family and was submitted to a positive selection pressure in the hominidae clade. Some residues under positive selection are of great importance for the normal function of the protein and thus for female fertility. Y235 represents a critical residue in the determination of BMP15 biological activity, thus indirectly confirming its role in the onset of POI in women.
Rational design of human DNA ligase inhibitors that target cellular DNA replication and repair.[Pubmed:18451142]
Cancer Res. 2008 May 1;68(9):3169-77.
Based on the crystal structure of human DNA ligase I complexed with nicked DNA, computer-aided drug design was used to identify compounds in a database of 1.5 million commercially available low molecular weight chemicals that were predicted to bind to a DNA-binding pocket within the DNA-binding domain of DNA ligase I, thereby inhibiting DNA joining. Ten of 192 candidates specifically inhibited purified human DNA ligase I. Notably, a subset of these compounds was also active against the other human DNA ligases. Three compounds that differed in their specificity for the three human DNA ligases were analyzed further. L82 inhibited DNA ligase I, L67 inhibited DNA ligases I and III, and L189 inhibited DNA ligases I, III, and IV in DNA joining assays with purified proteins and in cell extract assays of DNA replication, base excision repair, and nonhomologous end-joining. L67 and L189 are simple competitive inhibitors with respect to nicked DNA, whereas L82 is an uncompetitive inhibitor that stabilized complex formation between DNA ligase I and nicked DNA. In cell culture assays, L82 was cytostatic whereas L67 and L189 were cytotoxic. Concordant with their ability to inhibit DNA repair in vitro, subtoxic concentrations of L67 and L189 significantly increased the cytotoxicity of DNA-damaging agents. Interestingly, the ligase inhibitors specifically sensitized cancer cells to DNA damage. Thus, these novel human DNA ligase inhibitors will not only provide insights into the cellular function of these enzymes but also serve as lead compounds for the development of anticancer agents.
Mutation of dileucine-like motifs in the human immunodeficiency virus type 1 capsid disrupts virus assembly, gag-gag interactions, gag-membrane binding, and virion maturation.[Pubmed:16873251]
J Virol. 2006 Aug;80(16):7939-51.
The human immunodeficiency virus type 1 (HIV-1) Gag precursor protein Pr55(Gag) drives the assembly and release of virus-like particles in the infected cell. The capsid (CA) domain of Gag plays an important role in these processes by promoting Gag-Gag interactions during assembly. The C-terminal domain (CTD) of CA contains two dileucine-like motifs (L189/L190 and I201/L202) implicated in regulating the localization of Gag to multivesicular bodies (MVBs). These dileucine-like motifs are located in the vicinity of the CTD dimer interface, a region of CA critical for Gag-Gag interactions during virus assembly and CA-CA interactions during core formation. To study the importance of the CA dileucine-like motifs in various aspects of HIV-1 replication, we introduced a series of mutations into these motifs in the context of a full-length, infectious HIV-1 molecular clone. CA mutants LL189,190AA and IL201,202AA were both severely impaired in virus particle production because of a variety of defects in the binding of Gag to membrane, Gag multimerization, and CA folding. In contrast to the model suggesting that the CA dileucine-like motifs regulate MVB targeting, the IL201,202AA mutation did not alter Gag localization to the MVB in either HeLa cells or macrophages. Revertants of single-amino-acid substitution mutants were obtained that no longer contained dileucine-like motifs but were nevertheless fully replication competent. The varied phenotypes of the mutants reported here provide novel insights into the interplay among Gag multimerization, membrane binding, virus assembly, CA dimerization, particle maturation, and virion infectivity.
Short-lived chlorine-36 in a Ca- and Al-rich inclusion from the Ningqiang carbonaceous chondrite.[Pubmed:15671168]
Proc Natl Acad Sci U S A. 2005 Feb 1;102(5):1306-11.
Excesses of sulfur-36 in sodalite, a chlorine-rich mineral, in a calcium- and aluminum-rich inclusion from the Ningqiang carbonaceous chondrite linearly correlate with chorine/sulfur ratios, providing direct evidence for the presence of short-lived chlorine-36 (with a half-life of 0.3 million years) in the early solar system. The best inferred (36Cl/35Cl)o ratios of the sodalite are approximately 5 x 10(-6). Different from other short-lived radionuclides, chlorine-36 was introduced into the inclusion by solid-gas reaction during secondary alteration. The alteration reaction probably took place at least 1.5 million years after the first formation of the inclusion, based on the correlated study of the 26Al-26Mg systems of the relict primary minerals and the alteration assemblages, from which we inferred an initial ratio of (36Cl/35Cl)o > or = 1.6 x 10(-4) at the time when calcium- and aluminum-rich inclusions formed. This discovery supports a supernova origin of short-lived nuclides [Cameron, A. G. W., Hoeflich, P., Myers, P. C. & Clayton, D. D. (1995) Astrophys. J. 447, L53; Wasserburg, G. J., Gallino, R. & Busso, M. (1998) Astrophys. J. 500, L189-L193], but presents a serious challenge for local irradiation models [Shu, F. H., Shang, H., Glassgold, A. E. & Lee, T. (1997) Science 277, 1475-1479; Gounelle, M., Shu, F. H., Shang, H., Glassgold, A. E., Rehm, K. E. & Lee, T. (2001) Astrophys. J. 548, 1051-1070]. Furthermore, the short-lived 36Cl may serve as a unique fine-scale chronometer for volatile-rock interaction in the early solar system because of its close association with aqueous and/or anhydrous alteration processes.
Crystallographic structure of the nuclease domain of 3'hExo, a DEDDh family member, bound to rAMP.[Pubmed:15451662]
J Mol Biol. 2004 Oct 15;343(2):305-12.
A human 3'-5'-exoribonuclease (3'hExo) has recently been identified and shown to be responsible for histone mRNA degradation. Functionally, 3'hExo and a stem-loop binding protein (SLBP) target opposite faces of a unique highly conserved stem-loop RNA scaffold towards the 3' end of histone mRNA, which is composed of a 6 bp stem and a 4 nt loop, followed by an ACCCA sequence. Its Caenorhabditis elegans homologue, ERI-1, has been shown to degrade small interfering RNA in vitro and to function as a negative regulator of RNA interference in neuronal cells. We have determined the structure of the nuclease domain (Nuc) of 3'hExo complexed with rAMP in the presence of Mg2+ at 1.6 A resolution. The Nuc domain adopts an alpha/beta globular fold, with four acidic residues coordinating a binuclear metal cluster within the active site, whose topology is related to DEDDh exonuclease family members, despite a very low level of primary sequence identity. The two magnesium cations in the Nuc active site are coordinated to D134, E136, D234 and D298, and together with H293, which can potentially act as a general base, provide a platform for hydrolytic cleavage of bound RNA in the 3' --> 5' direction. The bound rAMP is positioned within a deep active-site pocket, with its purine ring close-packed with the hydrophobic F185 and L189 side-chains and its sugar 2'-OH and 3'-OH groups hydrogen bonded to backbone atoms of Nuc. There are striking similarities between the active sites of Nuc and epsilon186, an Escherichia coli DNA polymerase III proofreading domain, providing a common hydrolytic cleavage mechanism for RNA degradation and DNA editing, respectively.
Characterization of the UDP-N-acetylgalactosamine binding domain of bovine polypeptide alphaN-acetylgalactosaminyltransferase T1.[Pubmed:15377782]
Protein Eng Des Sel. 2004 Aug;17(8):635-46.
UDP-GalNAc:polypeptide alphaN-acetylgalactosaminyltransferases (ppGaNTases) transfer GalNAc from UDP-GalNAc to Ser or Thr. Structural features underlying their enzymatic activity and their specificity are still unidentified. In order to get some insight into the donor substrate recognition, we used a molecular modelling approach on a portion of the catalytic site of the bovine ppGaNTase-T1. Fold recognition methods identified as appropriate templates the bovine alpha1,3galactosyltransferase and the human alpha1,3N-acetylgalactosaminyltransferase. A model of the ppGaNTase-T1 nucleotide-sugar binding site was built into which the UDP-GalNAc and the Mn2+ cation were docked. UDP-GalNAc fits best in a conformation where the GalNAc is folded back under the phosphates and is maintained in that special conformation through hydrogen bonds with R193. The ribose is found in van der Waals contacts with F124 and L189. The uracil is involved in a stacking interaction with W129 and forms a hydrogen bond with N126. The Mn2+ is found in coordination both with the phosphates of UDP and the DXH motif of the enzyme. Amino acids in contact with UDP-GalNAc in the model have been mutated and the corresponding soluble forms of the enzyme expressed in yeast. Their kinetic constants confirm the importance of these amino acids in donor substrate interactions.
Mutational analysis of apolipoprotein B mRNA editing enzyme (APOBEC1). structure-function relationships of RNA editing and dimerization.[Pubmed:10191286]
J Lipid Res. 1999 Apr;40(4):623-35.
APOBEC1 is the catalytic subunit of an enzyme complex that mediates apolipoprotein (apo) B mRNA editing. It dimerizes in vitro and requires complementation factor(s) for its editing activity. We have performed a systematic analysis of the structure-functional relationship of APOBEC1 by targeted mutagenesis of various sequence motifs within the protein. Using in vitro RNA editing assay, we found that basic amino acid clusters at the amino-terminal region R15R16R17 and R33K34, are essential for apoB mRNA editing. Mutation of R15R16R17 to K15K16K17 and mutation of R33K34 simultaneously to A33A34 almost completely abolished in vitro editing activity. The carboxy-terminal region of APOBEC1 contains a leucine-rich motif. Deletion analysis of this region indicates that residues 181 to 210 are important for in vitro apoB mRNA editing. Single amino acid substitutions demonstrate that L182, I185, and L189 are important residues required for normal editing function. Furthermore, the double mutant P190A/P191A also lost >90% of editing activity which suggests that a beta turn in this region of the molecule may be essential for proper functioning of APOBEC1. It was suggested that dimerization of APOBEC1 creates an active structure for deamination of apoB mRNA. When we examined the dimerization potential of truncated APOBEC1s using both amino and carboxy termini deletion mutants, we found that amino-terminal deletions up to residue A117 did not impair dimerization activity whereas carboxy-terminal deletions showed diminished dimerization. The systematic and extensive mutagenesis experiments in this study provide information on the role of various sequence motifs identified in APOBEC1 in enzyme catalysis and dimerization.


