(±)-BI-DCAS# 1416258-16-6 |
- Ro3280
Catalog No.:BCC3962
CAS No.:1062243-51-9
- MLN0905
Catalog No.:BCC3961
CAS No.:1228960-69-7
- Rigosertib
Catalog No.:BCC4296
CAS No.:592542-59-1
- Rigosertib sodium
Catalog No.:BCC4067
CAS No.:592542-60-4
- GSK461364
Catalog No.:BCC3788
CAS No.:929095-18-1
Quality Control & MSDS
3D structure
Package In Stock
Number of papers citing our products

| Cas No. | 1416258-16-6 | SDF | Download SDF |
| PubChem ID | 122173021 | Appearance | Powder |
| Formula | C25H27NO4 | M.Wt | 405.49 |
| Type of Compound | N/A | Storage | Desiccate at -20°C |
| Solubility | DMSO : ≥ 100 mg/mL (246.62 mM) *"≥" means soluble, but saturation unknown. | ||
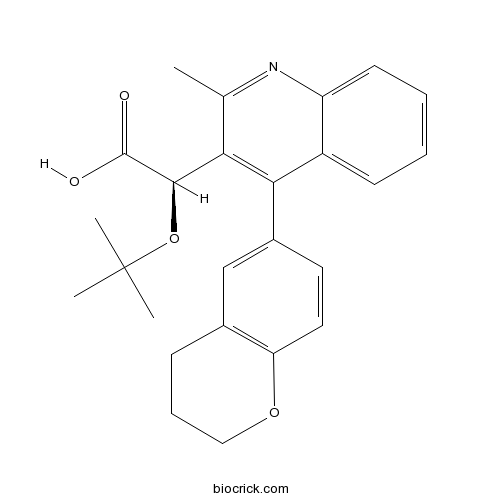
| Chemical Name | (2R)-2-[4-(3,4-dihydro-2H-chromen-6-yl)-2-methylquinolin-3-yl]-2-[(2-methylpropan-2-yl)oxy]acetic acid | ||
| SMILES | CC1=NC2=CC=CC=C2C(=C1C(C(=O)O)OC(C)(C)C)C3=CC4=C(C=C3)OCCC4 | ||
| Standard InChIKey | ZFERZAMPQIXCPM-HSZRJFAPSA-N | ||
| Standard InChI | InChI=1S/C25H27NO4/c1-15-21(23(24(27)28)30-25(2,3)4)22(18-9-5-6-10-19(18)26-15)17-11-12-20-16(14-17)8-7-13-29-20/h5-6,9-12,14,23H,7-8,13H2,1-4H3,(H,27,28)/t23-/m1/s1 | ||
| General tips | For obtaining a higher solubility , please warm the tube at 37 ℃ and shake it in the ultrasonic bath for a while.Stock solution can be stored below -20℃ for several months. We recommend that you prepare and use the solution on the same day. However, if the test schedule requires, the stock solutions can be prepared in advance, and the stock solution must be sealed and stored below -20℃. In general, the stock solution can be kept for several months. Before use, we recommend that you leave the vial at room temperature for at least an hour before opening it. |
||
| About Packaging | 1. The packaging of the product may be reversed during transportation, cause the high purity compounds to adhere to the neck or cap of the vial.Take the vail out of its packaging and shake gently until the compounds fall to the bottom of the vial. 2. For liquid products, please centrifuge at 500xg to gather the liquid to the bottom of the vial. 3. Try to avoid loss or contamination during the experiment. |
||
| Shipping Condition | Packaging according to customer requirements(5mg, 10mg, 20mg and more). Ship via FedEx, DHL, UPS, EMS or other couriers with RT, or blue ice upon request. | ||

(±)-BI-D Dilution Calculator

(±)-BI-D Molarity Calculator
| 1 mg | 5 mg | 10 mg | 20 mg | 25 mg | |
| 1 mM | 2.4662 mL | 12.3308 mL | 24.6615 mL | 49.323 mL | 61.6538 mL |
| 5 mM | 0.4932 mL | 2.4662 mL | 4.9323 mL | 9.8646 mL | 12.3308 mL |
| 10 mM | 0.2466 mL | 1.2331 mL | 2.4662 mL | 4.9323 mL | 6.1654 mL |
| 50 mM | 0.0493 mL | 0.2466 mL | 0.4932 mL | 0.9865 mL | 1.2331 mL |
| 100 mM | 0.0247 mL | 0.1233 mL | 0.2466 mL | 0.4932 mL | 0.6165 mL |
| * Note: If you are in the process of experiment, it's necessary to make the dilution ratios of the samples. The dilution data above is only for reference. Normally, it's can get a better solubility within lower of Concentrations. | |||||

Calcutta University

University of Minnesota

University of Maryland School of Medicine

University of Illinois at Chicago

The Ohio State University

University of Zurich

Harvard University

Colorado State University

Auburn University

Yale University

Worcester Polytechnic Institute

Washington State University

Stanford University

University of Leipzig

Universidade da Beira Interior

The Institute of Cancer Research

Heidelberg University

University of Amsterdam

University of Auckland

TsingHua University

The University of Michigan

Miami University

DRURY University

Jilin University

Fudan University

Wuhan University

Sun Yat-sen University

Universite de Paris

Deemed University

Auckland University

The University of Tokyo

Korea University
(±)-BI-D is a potent ALLINI(An allosteric IN inhibitor) that binds integrase at the LEDGF/p75 binding site. IC50 value: 2.4–2.9 μM(HIV-Luc infection of WT and Hdgfrp2 KO cells) [1] Target: integrase inhibitor in vitro: Approximately 2.4–2.9 μM of BI-D was required to inhibit 50% of HIV-Luc infection of WT and Hdgfrp2 KO cells, while the IC50 decreased dramatically, to 160–200 nM, in Psip1 and double-KO cells [1].
References:
[1]. Wang H, et al. HRP2 determines the efficiency and specificity of HIV-1 integration in LEDGF/p75 knockout cells but does not contribute to the antiviral activity of a potent LEDGF/p75-binding site integrase inhibitor. Nucleic Acids Res. 2012 Dec;40(22):115
[2]. Fader LD, et al. Minimizing the Contribution of Enterohepatic Recirculation to Clearance in Rat for the NCINI Class of Inhibitors of HIV. ACS Med Chem Lett. 2014 Apr 16;5(6):711-6.
- Dronedarone HCl
Catalog No.:BCC4777
CAS No.:141625-93-6
- Beta-D-glucopyranosyl oleanolate
Catalog No.:BCN6530
CAS No.:14162-53-9
- JW 642
Catalog No.:BCC6324
CAS No.:1416133-89-5
- UNC1215
Catalog No.:BCC2023
CAS No.:1415800-43-9
- Angustin B
Catalog No.:BCN7652
CAS No.:1415795-51-5
- Angustin A
Catalog No.:BCN7651
CAS No.:1415795-50-4
- ST-836 hydrochloride
Catalog No.:BCC1969
CAS No.:1415564-68-9
- PF-543 Citrate
Catalog No.:BCC1855
CAS No.:1415562-83-2
- PF-543
Catalog No.:BCC1854
CAS No.:1415562-82-1
- Crizotinib hydrochloride
Catalog No.:BCC5306
CAS No.:1415560-69-8
- GSK256066 2,2,2-trifluoroacetic acid
Catalog No.:BCC1605
CAS No.:1415560-64-3
- CDK9 inhibitor
Catalog No.:BCC1465
CAS No.:1415559-43-1
- Dronedarone
Catalog No.:BCN2176
CAS No.:141626-36-0
- CU CPT 22
Catalog No.:BCC6320
CAS No.:1416324-85-0
- GR 94800
Catalog No.:BCC5799
CAS No.:141636-65-9
- Ivangustin
Catalog No.:BCN3507
CAS No.:14164-59-1
- Thrombin Receptor Activator for Peptide 5 (TRAP-5)
Catalog No.:BCC1025
CAS No.:141685-53-2
- NPEC-caged-D-AP5
Catalog No.:BCC7895
CAS No.:1416943-27-5
- Galanin (2-29) (rat)
Catalog No.:BCC5763
CAS No.:141696-11-9
- Faropenem daloxate
Catalog No.:BCC1571
CAS No.:141702-36-5
- RI-2
Catalog No.:BCC6424
CAS No.:1417162-36-7
- H-Phe(4-Cl)-OH
Catalog No.:BCC3171
CAS No.:14173-39-8
- H-DL-Phe(4-Cl)-OMe.HCl
Catalog No.:BCC3175
CAS No.:14173-40-1
- Exenatide acetate
Catalog No.:BCN8514
CAS No.:141732-76-5
The allosteric HIV-1 integrase inhibitor BI-D affects virion maturation but does not influence packaging of a functional RNA genome.[Pubmed:25072705]
PLoS One. 2014 Jul 29;9(7):e103552.
The viral integrase (IN) is an essential protein for HIV-1 replication. IN inserts the viral dsDNA into the host chromosome, thereby aided by the cellular co-factor LEDGF/p75. Recently a new class of integrase inhibitors was described: allosteric IN inhibitors (ALLINIs). Although designed to interfere with the IN-LEDGF/p75 interaction to block HIV DNA integration during the early phase of HIV-1 replication, the major impact was surprisingly found on the process of virus maturation during the late phase, causing a reverse transcription defect upon infection of target cells. Virus particles produced in the presence of an ALLINI are misformed with the ribonucleoprotein located outside the virus core. Virus assembly and maturation are highly orchestrated and regulated processes in which several viral proteins and RNA molecules closely interact. It is therefore of interest to study whether ALLINIs have unpredicted pleiotropic effects on these RNA-related processes. We confirm that the ALLINI BI-D inhibits virus replication and that the produced virus is non-infectious. Furthermore, we show that the wild-type level of HIV-1 genomic RNA is packaged in virions and these genomes are in a dimeric state. The tRNAlys3 primer for reverse transcription was properly placed on this genomic RNA and could be extended ex vivo. In addition, the packaged reverse transcriptase enzyme was fully active when extracted from virions. As the RNA and enzyme components for reverse transcription are properly present in virions produced in the presence of BI-D, the inhibition of reverse transcription is likely to reflect the mislocalization of the components in the aberrant virus particle.
The Competitive Interplay between Allosteric HIV-1 Integrase Inhibitor BI/D and LEDGF/p75 during the Early Stage of HIV-1 Replication Adversely Affects Inhibitor Potency.[Pubmed:26910179]
ACS Chem Biol. 2016 May 20;11(5):1313-21.
Allosteric HIV-1 integrase inhibitors (ALLINIs) have recently emerged as a promising class of antiretroviral agents and are currently in clinical trials. In infected cells, ALLINIs potently inhibit viral replication by impairing virus particle maturation but surprisingly exhibit a reduced EC50 for inhibiting HIV-1 integration in target cells. To better understand the reduced antiviral activity of ALLINIs during the early stage of HIV-1 replication, we investigated the competitive interplay between a potent representative ALLINI, BI/D, and LEDGF/p75 with HIV-1 integrase. While the principal binding sites of BI/D and LEDGF/p75 overlap at the integrase catalytic core domain dimer interface, we show that the inhibitor and the cellular cofactor induce markedly different multimerization patterns of full-length integrase. LEDGF/p75 stabilizes an integrase tetramer through the additional interactions with the integrase N-terminal domain, whereas BI/D induces protein-protein interactions in C-terminal segments that lead to aberrant, higher-order integrase multimerization. We demonstrate that LEDGF/p75 binds HIV-1 integrase with significantly higher affinity than BI/D and that the cellular protein is able to reverse the inhibitor induced aberrant, higher-order integrase multimerization in a dose-dependent manner in vitro. Consistent with these observations, alterations of the cellular levels of LEDGF/p75 markedly affected BI/D EC50 values during the early steps of HIV-1 replication. Furthermore, genome-wide sequencing of HIV-1 integration sites in infected cells demonstrate that LEDGF/p75-dependent integration site selection is adversely affected by BI/D treatment. Taken together, our studies elucidate structural and mechanistic details of the interplay between LEDGF/p75 and BI/D during the early stage of HIV-1 replication.
Rejuvenating Bi(d)ology.[Pubmed:23069655]
Oncogene. 2013 Jul 4;32(27):3213-3219.
The BH3-only Bid protein is a critical sentinel of cellular stress in the liver and the hematopoietic system. Bid's initial 'claim to fame' came from its ability-as a caspase-truncated product-to trigger the mitochondrial apoptotic program following death receptor activation. Today we know that Bid can response to multiple types of proteases, which are activated under different conditions such as T-cell activation, ischemical reperfusion injury and lysosomal injury. Activation of the mitochondrial apoptotic program by Bid-via its recently identified receptor mitochondrial carrier homolog 2-involves multiple mechanisms, including release of cytochrome c and second mitochondria-derived activator of caspase (Smac), alteration of mitochondrial cristae organization, generation of reactive oxygen species and engagement of the permeability transition pore. Bid is also emerging-in its full-length form-as a pivotal sentinel of DNA damage in the bone marrow regulated by the ataxia telangiectasia mutated (ATM)/ataxia telangiectasia and Rad3-related (ATR) kinases. The ATM/ATR-Bid pathway is critically involved in preserving the quiescence and survival of hematopoietic stem cells both in the absence and presence of external stress, and a large part of this review will be dedicated to recent advances in this area of research.
The mechanism of H171T resistance reveals the importance of Ndelta-protonated His171 for the binding of allosteric inhibitor BI-D to HIV-1 integrase.[Pubmed:25421939]
Retrovirology. 2014 Nov 25;11:100.
BACKGROUND: Allosteric HIV-1 integrase (IN) inhibitors (ALLINIs) are an important new class of anti-HIV-1 agents. ALLINIs bind at the IN catalytic core domain (CCD) dimer interface occupying the principal binding pocket of its cellular cofactor LEDGF/p75. Consequently, ALLINIs inhibit HIV-1 IN interaction with LEDGF/p75 as well as promote aberrant IN multimerization. Selection of viral strains emerging under the inhibitor pressure has revealed mutations at the IN dimer interface near the inhibitor binding site. RESULTS: We have investigated the effects of one of the most prevalent substitutions, H171T IN, selected under increasing pressure of ALLINI BI-D. Virus containing the H171T IN substitution exhibited an ~68-fold resistance to BI-D treatment in infected cells. These results correlated with ~84-fold reduced affinity for BI-D binding to recombinant H171T IN CCD protein compared to its wild type (WT) counterpart. However, the H171T IN substitution only modestly affected IN-LEDGF/p75 binding and allowed HIV-1 containing this substitution to replicate at near WT levels. The x-ray crystal structures of BI-D binding to WT and H171T IN CCD dimers coupled with binding free energy calculations revealed the importance of the Ndelta- protonated imidazole group of His171 for hydrogen bonding to the BI-D tert-butoxy ether oxygen and establishing electrostatic interactions with the inhibitor carboxylic acid, whereas these interactions were compromised upon substitution to Thr171. CONCLUSIONS: Our findings reveal a distinct mechanism of resistance for the H171T IN mutation to ALLINI BI-D and indicate a previously undescribed role of the His171 side chain for binding the inhibitor.


