Methyl jasmonateCAS# 1211-29-6 |
Quality Control & MSDS
3D structure
Package In Stock
Number of papers citing our products

| Cas No. | 1211-29-6 | SDF | Download SDF |
| PubChem ID | 24208748.0 | Appearance | Powder |
| Formula | C13H20O3 | M.Wt | 224.3 |
| Type of Compound | N/A | Storage | Desiccate at -20°C |
| Solubility | Soluble in Chloroform,Dichloromethane,Ethyl Acetate,DMSO,Acetone,etc. | ||
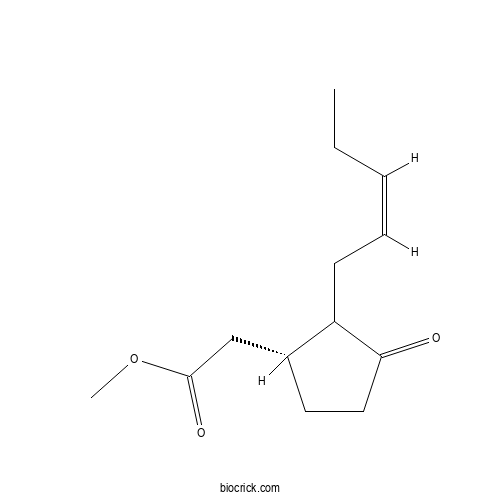
| Chemical Name | methyl 2-[(1R)-3-oxo-2-[(Z)-pent-2-enyl]cyclopentyl]acetate | ||
| SMILES | CCC=CCC1C(CCC1=O)CC(=O)OC | ||
| Standard InChIKey | GEWDNTWNSAZUDX-JXWYNLIVSA-N | ||
| Standard InChI | InChI=1S/C13H20O3/c1-3-4-5-6-11-10(7-8-12(11)14)9-13(15)16-2/h4-5,10-11H,3,6-9H2,1-2H3/b5-4-/t10-,11?/m1/s1 | ||
| General tips | For obtaining a higher solubility , please warm the tube at 37 ℃ and shake it in the ultrasonic bath for a while.Stock solution can be stored below -20℃ for several months. We recommend that you prepare and use the solution on the same day. However, if the test schedule requires, the stock solutions can be prepared in advance, and the stock solution must be sealed and stored below -20℃. In general, the stock solution can be kept for several months. Before use, we recommend that you leave the vial at room temperature for at least an hour before opening it. |
||
| About Packaging | 1. The packaging of the product may be reversed during transportation, cause the high purity compounds to adhere to the neck or cap of the vial.Take the vail out of its packaging and shake gently until the compounds fall to the bottom of the vial. 2. For liquid products, please centrifuge at 500xg to gather the liquid to the bottom of the vial. 3. Try to avoid loss or contamination during the experiment. |
||
| Shipping Condition | Packaging according to customer requirements(5mg, 10mg, 20mg and more). Ship via FedEx, DHL, UPS, EMS or other couriers with RT, or blue ice upon request. | ||

Methyl jasmonate Dilution Calculator

Methyl jasmonate Molarity Calculator
| 1 mg | 5 mg | 10 mg | 20 mg | 25 mg | |
| 1 mM | 4.4583 mL | 22.2916 mL | 44.5831 mL | 89.1663 mL | 111.4579 mL |
| 5 mM | 0.8917 mL | 4.4583 mL | 8.9166 mL | 17.8333 mL | 22.2916 mL |
| 10 mM | 0.4458 mL | 2.2292 mL | 4.4583 mL | 8.9166 mL | 11.1458 mL |
| 50 mM | 0.0892 mL | 0.4458 mL | 0.8917 mL | 1.7833 mL | 2.2292 mL |
| 100 mM | 0.0446 mL | 0.2229 mL | 0.4458 mL | 0.8917 mL | 1.1146 mL |
| * Note: If you are in the process of experiment, it's necessary to make the dilution ratios of the samples. The dilution data above is only for reference. Normally, it's can get a better solubility within lower of Concentrations. | |||||

Calcutta University

University of Minnesota

University of Maryland School of Medicine

University of Illinois at Chicago

The Ohio State University

University of Zurich

Harvard University

Colorado State University

Auburn University

Yale University

Worcester Polytechnic Institute

Washington State University

Stanford University

University of Leipzig

Universidade da Beira Interior

The Institute of Cancer Research

Heidelberg University

University of Amsterdam

University of Auckland

TsingHua University

The University of Michigan

Miami University

DRURY University

Jilin University

Fudan University

Wuhan University

Sun Yat-sen University

Universite de Paris

Deemed University

Auckland University

The University of Tokyo

Korea University
- Ginsenoside Ra6
Catalog No.:BCX0705
CAS No.:1346522-89-1
- Endothalic acid
Catalog No.:BCX0704
CAS No.:145-73-3
- Epirosmanol
Catalog No.:BCX0703
CAS No.:93380-12-2
- Mussaenosidic acid
Catalog No.:BCX0702
CAS No.:82451-22-7
- Methylcantharidinimide
Catalog No.:BCX0701
CAS No.:76970-78-0
- Cauloside D
Catalog No.:BCX0700
CAS No.:12672-45-6
- Cyclanoline
Catalog No.:BCX0699
CAS No.:18556-27-9
- Myricetin 3-O-rutinoside
Catalog No.:BCX0698
CAS No.:41093-68-9
- Desacylsenegasaponin B
Catalog No.:BCX0697
CAS No.:163589-51-3
- Quercetin 3-O-[beta-D-xylosyl-(1->2)-beta-D-glucoside]
Catalog No.:BCX0696
CAS No.:83048-35-5
- Cavidine
Catalog No.:BCX0695
CAS No.:32728-75-9
- Polygalasaponin XXVIII
Catalog No.:BCX0694
CAS No.:176182-01-7
- 5-Hydroxy-2′,3,4′,7-tetramethoxyflavone
Catalog No.:BCX0707
CAS No.:19056-75-8
- Oroxylin A-7-glucoside
Catalog No.:BCX0708
CAS No.:36948-77-3
- Oleandrigenin
Catalog No.:BCX0709
CAS No.:465-15-6
- Digitoxigenin
Catalog No.:BCX0710
CAS No.:143-62-4
- Pinolenic acid
Catalog No.:BCX0711
CAS No.:16833-54-8
- Oleandrin,anhydro-16-deacetyl-
Catalog No.:BCX0712
CAS No.:69549-58-2
- Epigallocatechin gallate octaacetate
Catalog No.:BCX0713
CAS No.:148707-39-5
- Bufotenidine
Catalog No.:BCX0714
CAS No.:487-91-2
- Salsolinol
Catalog No.:BCX0715
CAS No.:27740-96-1
- Prunetrin
Catalog No.:BCX0716
CAS No.:154-36-9
- limocitrin -3-O-rutinoside
Catalog No.:BCX0717
CAS No.:79384-27-3
- Anthrone
Catalog No.:BCX0718
CAS No.:90-44-8
RhMED15a-like, a subunit of the Mediator complex, is involved in the drought stress response in Rosa hybrida.[Pubmed:38684962]
BMC Plant Biol. 2024 Apr 30;24(1):351.
BACKGROUND: Rose (Rosa hybrida) is a globally recognized ornamental plant whose growth and distribution are strongly limited by drought stress. The role of Mediator, a multiprotein complex crucial for RNA polymerase II-driven transcription, has been elucidated in drought stress responses in plants. However, its physiological function and regulatory mechanism in horticultural crop species remain elusive. RESULTS: In this study, we identified a Tail module subunit of Mediator, RhMED15a-like, in rose. Drought stress, as well as treatment with Methyl jasmonate (MeJA) and abscisic acid (ABA), significantly suppressed the transcript level of RhMED15a-like. Overexpressing RhMED15a-like markedly bolstered the osmotic stress tolerance of Arabidopsis, as evidenced by increased germination rate, root length, and fresh weight. In contrast, the silencing of RhMED15a-like through virus induced gene silencing in rose resulted in elevated malondialdehyde accumulation, exacerbated leaf wilting, reduced survival rate, and downregulated expression of drought-responsive genes during drought stress. Additionally, using RNA-seq, we identified 972 differentially expressed genes (DEGs) between tobacco rattle virus (TRV)-RhMED15a-like plants and TRV controls. Gene Ontology (GO) analysis revealed that some DEGs were predominantly associated with terms related to the oxidative stress response, such as 'response to reactive oxygen species' and 'peroxisome'. Furthermore, Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment highlighted pathways related to 'plant hormone signal transduction', in which the majority of DEGs in the jasmonate (JA) and ABA signalling pathways were induced in TRV-RhMED15a-like plants. CONCLUSION: Our findings underscore the pivotal role of the Mediator subunit RhMED15a-like in the ability of rose to withstand drought stress, probably by controlling the transcript levels of drought-responsive genes and signalling pathway elements of stress-related hormones, providing a solid foundation for future research into the molecular mechanisms underlying drought tolerance in rose.
Methyl jasmonate ameliorates pain-induced learning and memory impairments through regulating the expression of genes involved in neuroinflammation.[Pubmed:38680072]
Brain Behav. 2024 May;14(5):e3502.
OBJECTIVE: Orofacial pain with high prevalence is one of the substantial human health issues. The importance of this matter became more apparent when it was revealed that orofacial pain, directly and indirectly, affects cognition performances. Currently, researchers have focused on investigating pharmaceutics to alleviate pain and ameliorate its subsequent cognitive impairments. DESIGN: In this study, the rats were first treated with the central administration of Methyl jasmonate (MeJA), which is an antioxidant and anti-inflammatory bio-compound. After 20 min, orofacial pain was induced in the rats by the injection of capsaicin in their dental pulp. Subsequently, the animals' pain behaviors were analyzed, and the effects of pain and MeJA treatments on rats learning and memory were evaluated/compared using the Morris water maze (MWM) test. In addition, the expression of tumor necrosis factor-alpha (TNF-alpha), IL-1beta, BDNF, and COX-2 genes in the rats' hippocampus was evaluated using real-time polymerase chain reaction. RESULTS: Experiencing orofacial pain resulted in a significant decline in the rats learning and memory. However, the central administration of 20 mug/rat of MeJA effectively mitigated these impairments. In the MWM, the performance of the MeJA-treated rats showed a two- to threefold improvement compared to the nontreated ones. Moreover, in the hippocampus of pain-induced rats, the expression of pro-inflammatory factors TNF-alpha, IL-1beta, and COX-2 significantly increased, whereas the BDNF expression decreased. In contrast, MeJA downregulated the pro-inflammatory factors and upregulated the BDNF by more than 50%. CONCLUSIONS: These findings highlight the notable antinociceptive potential of MeJA and its ability to inhibit pain-induced learning and memory dysfunction through its anti-inflammatory effect.
Identification and Expression Analysis of the WOX Transcription Factor Family in Foxtail Millet (Setaria italica L.).[Pubmed:38674410]
Genes (Basel). 2024 Apr 10;15(4):476.
WUSCHEL-related homeobox (WOX) transcription factors are unique to plants and play pivotal roles in plant development and stress responses. In this investigation, we acquired protein sequences of foxtail millet WOX gene family members through homologous sequence alignment and a hidden Markov model (HMM) search. Utilizing conserved domain prediction, we identified 13 foxtail millet WOX genes, which were classified into ancient, intermediate, and modern clades. Multiple sequence alignment results revealed that all WOX proteins possess a homeodomain (HD). The SiWOX genes, clustered together in the phylogenetic tree, exhibited analogous protein spatial structures, gene structures, and conserved motifs. The foxtail millet WOX genes are distributed across 7 chromosomes, featuring 3 pairs of tandem repeats: SiWOX1 and SiWOX13, SiWOX4 and SiWOX5, and SiWOX11 and SiWOX12. Collinearity analysis demonstrated that WOX genes in foxtail millet exhibit the highest collinearity with green foxtail, followed by maize. The SiWOX genes primarily harbor two categories of cis-acting regulatory elements: Stress response and plant hormone response. Notably, prominent hormones triggering responses include Methyl jasmonate, abscisic acid, gibberellin, auxin, and salicylic acid. Analysis of SiWOX expression patterns and hormone responses unveiled potential functional diversity among different SiWOX genes in foxtail millet. These findings lay a solid foundation for further elucidating the functions and evolution of SiWOX genes.
Isolation, Characterization, and Expression Analysis of NAC Transcription Factor from Andrographis paniculata (Burm. f.) Nees and Their Role in Andrographolide Production.[Pubmed:38674357]
Genes (Basel). 2024 Mar 28;15(4):422.
Andrographis paniculata (Burm. f.) Nees is an important medicinal plant known for its bioactive compound andrographolide. NAC transcription factors (NAM, ATAF1/2, and CUC2) play a crucial role in secondary metabolite production, stress responses, and plant development through hormonal signaling. In this study, a putative partial transcript of three NAC family genes (ApNAC83, ApNAC21 22 and ApNAC02) was used to isolate full length genes using RACE. Bioinformatics analyses such as protein structure prediction, cis-acting regulatory elements, and gene ontology analysis were performed. Based on in silico predictions, the diterpenoid profiling of the plant's leaves (five-week-old) and the real-time PCR-based expression analysis of isolated NAC genes under abscisic acid (ABA) treatment were performed. Additionally, the expression analysis of isolated NAC genes under MeJA treatment and transient expression in Nicotiana tabacum was performed. Full-length sequences of three members of the NAC transcription factor family, ApNAC83 (1102 bp), ApNAC21 22 (996 bp), and ApNAC02 (1011 bp), were isolated and subjected to the promoter and gene ontology analysis, which indicated their role in transcriptional regulation, DNA binding, ABA-activated signaling, and stress management. It was observed that ABA treatment leads to a higher accumulation of andrographolide and 14-deoxyandrographolide content, along with the upregulation of ApNAC02 (9.6-fold) and the downregulation of ApNAC83 and ApNAC21 22 in the leaves. With Methyl jasmonate treatment, ApNAC21 22 expression decreased, while ApNAC02 increased (1.9-fold), with no significant change being observed in ApNAC83. The transient expression of the isolated NAC genes in a heterologous system (Nicotiana benthamiana) demonstrated their functional transcriptional activity, leading to the upregulation of the NtHMGR gene, which is related to the terpene pathway in tobacco. The expression analysis and heterologous expression of ApNAC21 22 and ApNAC02 indicated their role in andrographolide biosynthesis.
Genome-wide identification and analysis of DEAD-box RNA helicases in Gossypium hirsutum.[Pubmed:38663690]
Gene. 2024 Apr 23:148495.
DEAD-box RNA helicases, a prominent subfamily within the RNA helicase superfamily 2 (SF2), play crucial roles in the growth, development, and abiotic stress responses of plants. This study identifies 146 DEAD-box RNA helicase genes (GhDEADs) and categorizes them into four Clades (Clade A-D) through phylogenetic analysis. Promoter analysis reveals cis-acting elements linked to plant responses to light, Methyl jasmonate (MeJA), abscisic acid (ABA), low temperature, and drought. RNA-seq data demonstrate that Clade C GhDEADs exhibit elevated and ubiquitous expression across different tissues, validating their connection to leaf development through real-time quantitative polymerase chain reaction (RT-qPCR) analysis. Notably, over half of GhDEADs display up-regulation in the leaves of virus-induced gene silencing (VIGS) plants of GhVIR-A/D (members of m(6)A methyltransferase complex, which regulate leaf morphogenesis). In conclusion, this study offers a comprehensive insight into GhDEADs, emphasizing their potential involvement in leaf development.
[Characterization of sequences, expression profiling, and natural allelic variation analysis of the MYC gene family in sorghum (Sorghum bicolor)].[Pubmed:38658156]
Sheng Wu Gong Cheng Xue Bao. 2024 Apr 25;40(4):1170-1194.
Sorghum aphid (Melanaphis sacchari) and head smut fungi (Sporisorium reilianum) infesting sorghum cause delayed growth and development, and reduce yield and quality. This study use bioinformatics and molecular biological approaches to profile the gene expression pattern during sorghum development and under pest infestation, and analyzed the natural allelic DNA variation of sorghum MYC gene family. The findings provide insights for potential application in breeding the stress resistant and high productivity sorghum varieties. The results indicated that there are 28 MYC genes identified in sorghum genome, distributed on 10 chromosomes. The bHLH_MYC_N and HLH domains are the conserved domains of the MYC gene in sorghum. Gene expression analysis showed that SbbHLH35.7g exhibited high expression levels in leaves, SbAbaIn showed strong expression in early grains, and SbMYC2.1g showed high expression levels in mature pollen. In anti-aphid strains at the 5-leaf stage, SbAbaIn, SbLHW.4g and SbLHW.2g were significantly induced in leaves, while SbbHLH35.7g displayed the highest expression level in panicle tissue, which was significantly induced by the infection of head smut. Promoter cis-element analysis identified Methyl jasmonate (MJ), abscisic acid (ABA), salicylic acid (SA) and MYB-binding sites related to drought-stress inducibility. Furthermore, genomic resequencing data analysis revealed natural allelic DNA variations such as single nucleotide polymorphism (SNP) and insertion-deletion (INDEL) for the key SbMYCs. Protein interaction network analysis using STRING indicated that SbAbaIn interacts with TIFYdomain protein, and SbbHLH35.7g interacts with MDR and imporin. SbMYCs exhibited temporal and spatial expression patterns and played vital roles during the sorghum development. Infestation by sugarcane aphids and head smut fungi induced the expression of SbAbaIn and SbbHLH35.7g, respectively. SbAbaIn modulated the jasmonic acid (JA) pathway to regulate the expression of defensive genes, conferring resistance to insects. On the other hand, SbbHLH35.7g participated in detoxification reactions to defend against pathogens.
Changes in Physiological Traits, Gene Expression and Phytochemical Profile of Mentha piperita in Response to Elicitor.[Pubmed:38653889]
Biochem Genet. 2024 Apr 23.
Peppermint (Mentha piperita) is a perennial medicinal plant containing active ingredients that can be used for treating liver and prostate cancers, acute respiratory infections, allergies, digestive problems, neuralgia, and migraines. The objective of this research is to investigate the expression of essential genes in the menthol pathway of Mentha piperita, including Pulegone reductase (Pr), Menthofuran synthase (Mfs), and limonene synthase (Ls) using qPCR, physiological analysis and essential oil composition in response to Methyl jasmonate (MeJA) (0.5 mM) elicitation. Physiological analysis showed that 0.5 mM MeJA triggers defensive responsiveness in Mentha piperita by increasing superoxide dismutase (SOD) and Peroxidase (POD) enzymes activity. The highest transcript levels of Pr and Mfs genes were observed during 8 and 12 h after treatment respectively, but following 24 h, they were down-regulated. Essential oil analysis indicated that the percentage of constituents in the essential oil was changed using MeJA at 48 h and 96 h after post-treatment. Effective antimicrobial compounds, alpha-pinene, beta-pinene, linalool and methyl acetate, were induced after 48 h. A non-significant positive relationship was detected between menthol content, and expression of the Pr and Mfs genes. Due to the significant change in the expression of Pr and Mfs genes in the menthol pathway, role of Pr gene in directing the pathway to the valuable compound menthol and deviation of the menthol pathway to the menthofuran as an undesirable component of essential oil by Mfs gene, it can be deduced that they are the most critical genes in response to MeJA treatment, which are appropriate candidates for metabolite engineering. In addition, MeJA improved defensive responsiveness and percentage of some constituents with antimicrobial properties in Mentha piperita.
Transcriptomic data reveals the dynamics of terpenoids biosynthetic pathway of fenugreek.[Pubmed:38649807]
BMC Genomics. 2024 Apr 22;25(1):390.
Medicinal plants are rich sources for treating various diseases due their bioactive secondary metabolites. Fenugreek (Trigonella foenum-graecum) is one of the medicinal plants traditionally used in human nutrition and medicine which contains an active substance, called diosgenin, with anticancer properties. Biosynthesis of this important anticancer compound in fenugreek can be enhanced using eliciting agents which involves in manipulation of metabolite and biochemical pathways stimulating defense responses. Methyl jasmonate elicitor was used to increase diosgenin biosynthesis in fenugreek plants. However, the molecular mechanism and gene expression profiles underlying diosgening accumulation remain unexplored. In the current study we performed an extensive analysis of publicly available RNA-sequencing datasets to elucidate the biosynthesis and expression profile of fenugreek plants treated with Methyl jasmonate. For this purpose, seven read datasets of Methyl jasmonate treated plants were obtained that were covering several post-treatment time points (6-120 h). Transcriptomics analysis revealed upregulation of several key genes involved in diosgenein biosynthetic pathway including Squalene synthase (SQS) as the first committed step in diosgenin biosynthesis as well as Squalene Epoxidase (SEP) and Cycloartenol Synthase (CAS) upon Methyl jasmonate application. Bioinformatics analysis, including gene ontology enrichment and pathway analysis, further supported the involvement of these genes in diosgenin biosynthesis. The bioinformatics analysis led to a comprehensive validation, with expression profiling across three different fenugreek populations treated with the same Methyl jasmonate application. Initially, key genes like SQS, SEP, and CAS showed upregulation, followed by later upregulation of Delta24, suggesting dynamic pathway regulation. Real-time PCR confirmed consistent upregulation of SQS and SEP, peaking at 72 h. Additionally, candidate genes Delta24 and SMT1 highlighted roles in directing metabolic flux towards diosgenin biosynthesis. This integrated approach validates the bioinformatics findings and elucidates fenugreek's molecular response to Methyl jasmonate elicitation, offering insights for enhancing diosgenin yield. The assembled transcripts and gene expression profiles are deposited in the Zenodo open repository at https://doi.org/10.5281/zenodo.8155183 .
Genome-wide identification and evolutionary analysis of the NRAMP gene family in the AC genomes of Brassica species.[Pubmed:38649805]
BMC Plant Biol. 2024 Apr 23;24(1):311.
BACKGROUND: Brassica napus, a hybrid resulting from the crossing of Brassica rapa and Brassica oleracea, is one of the most important oil crops. Despite its significance, B. napus productivity faces substantial challenges due to heavy metal stress, especially in response to cadmium (Cd), which poses a significant threat among heavy metals. Natural resistance-associated macrophage proteins (NRAMPs) play pivotal roles in Cd uptake and transport within plants. However, our understanding of the role of BnNRAMPs in B. napus is limited. Thus, this study aimed to conduct genome-wide identification and bioinformatics analysis of three Brassica species: B. napus, B. rapa, and B. oleracea. RESULTS: A total of 37 NRAMPs were identified across the three Brassica species and classified into two distinct subfamilies based on evolutionary relationships. Conservative motif analysis revealed that motif 6 and motif 8 might significantly contribute to the differentiation between subfamily I and subfamily II within Brassica species. Evolutionary analyses and chromosome mapping revealed a reduction in the NRAMP gene family during B. napus evolutionary history, resulting in the loss of an orthologous gene derived from BoNRAMP3.2. Cis-acting element analysis suggested potential regulation of the NRAMP gene family by specific plant hormones, such as abscisic acid (ABA) and Methyl jasmonate (MeJA). However, gene expression pattern analyses under hormonal or stress treatments indicated limited responsiveness of the NRAMP gene family to these treatments, warranting further experimental validation. Under Cd stress in B. napus, expression pattern analysis of the NRAMP gene family revealed a decrease in the expression levels of most BnNRAMP genes with increasing Cd concentrations. Notably, BnNRAMP5.1/5.2 exhibited a unique response pattern, being stimulated at low Cd concentrations and inhibited at high Cd concentrations, suggesting potential response mechanisms distinct from those of other NRAMP genes. CONCLUSIONS: In summary, this study indicates complex molecular dynamics within the NRAMP gene family under Cd stress, suggesting potential applications in enhancing plant resilience, particularly against Cd. The findings also offer valuable insights for further understanding the functionality and regulatory mechanisms of the NRAMP gene family.
Increased paclitaxel recovery from Taxus baccata vascular stem cells using novel in situ product recovery approaches.[Pubmed:38647629]
Bioresour Bioprocess. 2023 Sep 29;10(1):68.
In this study, several approaches were tested to optimise the production and recovery of the widely used anticancer drug Taxol((R)) (paclitaxel) from culturable vascular stem cells (VSCs) of Taxus baccata, which is currently used as a successful cell line for paclitaxel production. An in situ product recovery (ISPR) technique was employed, which involved combining three commercial macro-porous resin beads (HP-20, XAD7HP and HP-2MG) with batch and semi-continuous cultivations of the T. baccata VSCs after adding Methyl jasmonate (Me-JA) as an elicitor. The optimal resin combination resulted in 234 +/- 23 mg of paclitaxel per kg of fresh-weight cells, indicating a 13-fold improved yield compared to the control (with no resins) in batch cultivation. This resin treatment was further studied to evaluate the resins' removal capacity of reactive oxygen species (ROS), which can cause poor cell growth or reduce product synthesis. It was observed that the ISPR cultivations had fourfold less intracellular ROS concentration than that of the control; thus, a reduced ROS concentration established by the resin contributed to increased paclitaxel yield, contrary to previous studies. These paclitaxel yields are the highest reported to date using VSCs, and this scalable production method could be applied for a diverse range of similar compounds utilising plant cell culture.
Identification of the lateral organ boundary domain gene family and its preservation by exogenous salicylic acid in Cerasus humilis.[Pubmed:38633270]
Physiol Mol Biol Plants. 2024 Mar;30(3):401-415.
The gene family known as the Lateral Organ Boundary Domain (LBD) is responsible for producing transcription factors unique to plants, which play a crucial role in controlling diverse biological activities, including their growth and development. This research focused on examining Cerasus humilis'ChLBD gene, owing to its significant ecological, economic, and nutritional benefits. Examining the ChLBD gene family's member count, physicochemical characteristics, phylogenetic evolution, gene configuration, and motif revealed 41 ChLBD gene family members spread across 8 chromosomes, with ChLBD gene's full-length coding sequences (CDSs) ranging from 327 to 1737 base pairs, and the protein sequence's length spanning 109 (ChLBD30)-579 (ChLBD35) amino acids. The molecular weights vary from 12.068 (ChLBD30) to 62.748 (ChLBD35) kDa, and the isoelectric points span from 4.74 (ChLBD20) to 9.19 (ChLBD3). Categorizing them into two evolutionary subfamilies: class I with 5 branches, class II with 2, the majority of genes with a single intron, and most members of the same subclade sharing comparable motif structures. The results of collinearity analysis showed that there were 3 pairs of tandem repeat genes and 12 pairs of fragment repeat genes in the Cerasus humilis genome, and in the interspecific collinearity analysis, the number of collinear gene pairs with apples belonging to the same family of Rosaceae was the highest. Examination of cis-acting elements revealed that Methyl jasmonate response elements stood out as the most abundant, extensively dispersed in the promoter areas of class 1 and class 2 ChLBD. Genetic transcript analysis revealed that during Cerasus humilis' growth and maturation, ChLBD developed varied control mechanisms, with ChLBD27 and ChLBD40 potentially playing a role in managing color alterations in fruit ripening. In addition, the quality of calcium fruit will be affected by the environment during transportation and storage, and it is particularly important to use appropriate means to preserve the fruit. The research used salicylic acid-treated Cerasus humilis as the research object and employed qRT-PCR to examine the expression of six ChLBD genes throughout storage. Variations in the expression of the ChLBD gene were observed when exposed to salicylic acid, indicating that salicylic acid could influence ChLBD gene expression during the storage of fruits. This study's findings lay the groundwork for additional research into the biological role of the LBD gene in Cerasus humilis. SUPPLEMENTARY INFORMATION: The online version contains supplementary material available at 10.1007/s12298-024-01438-5.
LaMYC7, a positive regulator of linalool and caryophyllene biosynthesis, confers plant resistance to Pseudomonas syringae.[Pubmed:38623075]
Hortic Res. 2024 Feb 6;11(4):uhae044.
Linalool and caryophyllene are the main monoterpene and sesquiterpene compounds in lavender; however, the genes regulating their biosynthesis still remain many unknowns. Here, we identified LaMYC7, a positive regulator of linalool and caryophyllene biosynthesis, confers plant resistance to Pseudomonas syringae. LaMYC7 was highly expressed in glandular trichomes, and LaMYC7 overexpression could significantly increase the linalool and caryophyllene contents and reduce susceptibility to P. syringae in Nicotiana. In addition, the linalool possessed antimicrobial activity against P. syringae growth and acted dose-dependently. Further analysis demonstrated that LaMYC7 directly bound to the promoter region of LaTPS76, which encodes the terpene synthase (TPS) for caryophyllene biosynthesis, and that LaTPS76 was highly expressed in glandular trichomes. Notably, the LaMYC7 promoter contained hormone and stress-responsive regulatory elements and responded to various treatments, including ultraviolet, low temperature, salt, drought, Methyl jasmonate, and P. syringae infection treatments. Under these treatments, the changes in the linalool and caryophyllene contents were similar to those in LaMYC7 transcript abundance. Based on the results, LaMYC7 could respond to P. syringae infection in addition to being involved in linalool and caryophyllene biosynthesis. Thus, the MYC transcription factor gene LaMYC7 can be used in the breeding of high-yielding linalool and caryophyllene lavender varieties with pathogen resistance.


