PR-619Deubiquitylating enzymes (DBUs) inhibitor CAS# 2645-32-1 |
- NSC 687852 (b-AP15)
Catalog No.:BCC2389
CAS No.:1009817-63-3
- TCID
Catalog No.:BCC4449
CAS No.:30675-13-9
- SJB2-043
Catalog No.:BCC1952
CAS No.:63388-44-3
- WP1130
Catalog No.:BCC3686
CAS No.:856243-80-6
- Vialinin A
Catalog No.:BCC2367
CAS No.:858134-23-3
Quality Control & MSDS
3D structure
Package In Stock
Number of papers citing our products

| Cas No. | 2645-32-1 | SDF | Download SDF |
| PubChem ID | 2817763 | Appearance | Powder |
| Formula | C7H5N5S2 | M.Wt | 223.28 |
| Type of Compound | N/A | Storage | Desiccate at -20°C |
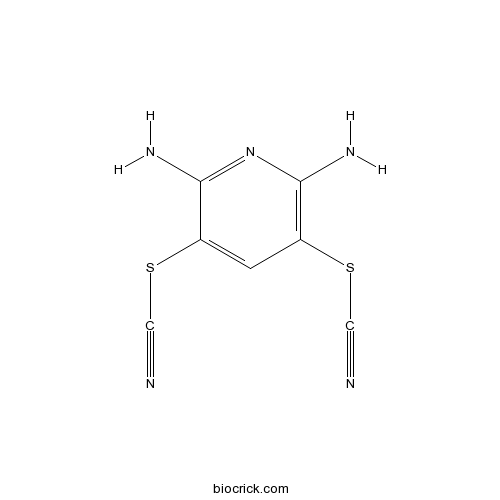
| Synonyms | 2,6-Diamino-3,5-dithiocyanopyridine | ||
| Solubility | DMSO : ≥ 21 mg/mL (94.05 mM) *"≥" means soluble, but saturation unknown. | ||
| Chemical Name | (2,6-diamino-5-thiocyanatopyridin-3-yl) thiocyanate | ||
| SMILES | C1=C(C(=NC(=C1SC#N)N)N)SC#N | ||
| Standard InChIKey | ZXOBLNBVNROVLC-UHFFFAOYSA-N | ||
| Standard InChI | InChI=1S/C7H5N5S2/c8-2-13-4-1-5(14-3-9)7(11)12-6(4)10/h1H,(H4,10,11,12) | ||
| General tips | For obtaining a higher solubility , please warm the tube at 37 ℃ and shake it in the ultrasonic bath for a while.Stock solution can be stored below -20℃ for several months. We recommend that you prepare and use the solution on the same day. However, if the test schedule requires, the stock solutions can be prepared in advance, and the stock solution must be sealed and stored below -20℃. In general, the stock solution can be kept for several months. Before use, we recommend that you leave the vial at room temperature for at least an hour before opening it. |
||
| About Packaging | 1. The packaging of the product may be reversed during transportation, cause the high purity compounds to adhere to the neck or cap of the vial.Take the vail out of its packaging and shake gently until the compounds fall to the bottom of the vial. 2. For liquid products, please centrifuge at 500xg to gather the liquid to the bottom of the vial. 3. Try to avoid loss or contamination during the experiment. |
||
| Shipping Condition | Packaging according to customer requirements(5mg, 10mg, 20mg and more). Ship via FedEx, DHL, UPS, EMS or other couriers with RT, or blue ice upon request. | ||
| Description | Broad spectrum, reversible DUB inhibitor. Exhibits limited activity against other proteases. Induces accumulation of polyubiquitinated proteins, but has no direct inhibitory effect on the proteasome. Cytotoxic in HEK293T and colorectal cancer cells. Stabilizes microtubule network in oligodendroglial cells. |

PR-619 Dilution Calculator

PR-619 Molarity Calculator
| 1 mg | 5 mg | 10 mg | 20 mg | 25 mg | |
| 1 mM | 4.4787 mL | 22.3934 mL | 44.7868 mL | 89.5736 mL | 111.967 mL |
| 5 mM | 0.8957 mL | 4.4787 mL | 8.9574 mL | 17.9147 mL | 22.3934 mL |
| 10 mM | 0.4479 mL | 2.2393 mL | 4.4787 mL | 8.9574 mL | 11.1967 mL |
| 50 mM | 0.0896 mL | 0.4479 mL | 0.8957 mL | 1.7915 mL | 2.2393 mL |
| 100 mM | 0.0448 mL | 0.2239 mL | 0.4479 mL | 0.8957 mL | 1.1197 mL |
| * Note: If you are in the process of experiment, it's necessary to make the dilution ratios of the samples. The dilution data above is only for reference. Normally, it's can get a better solubility within lower of Concentrations. | |||||

Calcutta University

University of Minnesota

University of Maryland School of Medicine

University of Illinois at Chicago

The Ohio State University

University of Zurich

Harvard University

Colorado State University

Auburn University

Yale University

Worcester Polytechnic Institute

Washington State University

Stanford University

University of Leipzig

Universidade da Beira Interior

The Institute of Cancer Research

Heidelberg University

University of Amsterdam

University of Auckland

TsingHua University

The University of Michigan

Miami University

DRURY University

Jilin University

Fudan University

Wuhan University

Sun Yat-sen University

Universite de Paris

Deemed University

Auckland University

The University of Tokyo

Korea University
(2,6-diamino-5-thiocyanatopyridin-3-yl) thiocyanate, also designated as PR-619, is a broad-range reversible and cell-permeable inhibitor of deubiquitylating enzyme (DUB)[1][2][3], potently suppresses the activity of almost all cysteine protease DUBs[4], but shows selectivity toward DUBs over other proteases, such as calpain 1, or cathepsins[3]. PR-619 induces (tumor) cell death with EC50 values in the low micromolar range [1].
Deubiquitylating enzyme (DUB), also called ubiquitin isopeptidase or deubiquitinating proteins, performs deubiquitination of target proteins. Ubiquitination, followed by proteasomal degradation, is a process of ubiquitin proteasome system (UPS). Failure of the ubiquitin proteasome system (UPS) and/or the autophagy pathway may result in aggregation of proteins, a pathological hallmark of many neurodegenerative diseases [2].
In OLN-t40 cells, 7-10 μM as the concentration range for PR-619 to exert its cytotoxicity was suggested, half maximal cytotoxicity was observed after a 24h-treatment with 9-10 μM PR-619. Similar to MG-132, PR-619 caused an increase in the abundance of ubiquitinated proteins within 24 h at the concentration range of 7-12.5μM. Tested with OLN-t40 cells, PR-619, unlike MG-132, did not inhibit the enzymatic activity of the proteasome in cellular lysates but only when taken up by living cells[2]. An in vitro system showed that PR-619 was able to stabilize the microtubule network and led to small tau aggregates surrounding the microtubule organizing center [5].
There is still not any available result regarding in vivo treatment in an animal body.
References:
[1]. Mikael Altun, Holger B. Kramer, Lianne I. Willems, et al. Activity-Based Chemical Proteomics Accelerates Inhibitor Development for Deubiquitylating Enzymes. Chemistry & Biology, 2011, 18(11): 1401-1412.
[2]. Veronika Seiberlicha, Olaf Goldbauma, Victoria Zhukareva, et al. The small molecule inhibitor PR-619 of deubiquitinating enzymes affects the microtubule network and causes protein aggregate formation in neural cells: Implications for neurodegenerative diseases. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, 2012, 1823(11): 2057–2068.
[3]. Iraia García-Santisteban, Godefridus J Peters, Elisa Giovannetti, et al. USP1 deubiquitinase: cellular functions, regulatory mechanisms and emerging potential as target in cancer therapy. Molecular Cancer, 2013, 12:91.
[4]. Maria Stella Ritorto, Richard Ewan, Ana B. Perez-Oliva, et al. Screening of DUB activity and specificity by MALDI-TOF mass spectrometry. Nature Communications, 2014, 5:4763.
[5]. Laura J Blair, Bo Zhang and Chad A Dickey, et al. Potential synergy between tau aggregation inhibitors and tau chaperone modulators. Alzheimer's Research & Therapy, 2013, 5:41.
- Bz-Arg-OEt.HCl
Catalog No.:BCC2686
CAS No.:2645-08-1
- Methyl 2,2-dithienylglycolate
Catalog No.:BCC9034
CAS No.:26447-85-8
- Oxypeucedanin hydrate
Catalog No.:BCN2698
CAS No.:2643-85-8
- 6',7'-Dihydroxybergamottin
Catalog No.:BCN5142
CAS No.:264234-05-1
- Methyl 5-{2-[(tert-butylamino)carbothioyl]carbohydrazonoyl}-1-(2,4-difluorophenyl)-1H-pyrazole-4-carboxylate
Catalog No.:BCC7906
CAS No.:264233-05-8
- SB 415286
Catalog No.:BCC3651
CAS No.:264218-23-7
- Z-Orn-OH
Catalog No.:BCC2757
CAS No.:2640-58-6
- DPPA (Kg)
Catalog No.:BCC2690
CAS No.:26386-88-9
- S 25585
Catalog No.:BCC7687
CAS No.:263849-50-9
- AG 045572
Catalog No.:BCC7464
CAS No.:263847-55-8
- 3-Acetoxy-27-hydroxy-20(29)-lupen-28-oic acid methyl ester
Catalog No.:BCN1466
CAS No.:263844-80-0
- 3,27-Dihydroxy-20(29)-lupen-28-oic acid methyl ester
Catalog No.:BCN1467
CAS No.:263844-79-7
- SSR 146977 hydrochloride
Catalog No.:BCC7635
CAS No.:264618-38-4
- MRS 1706
Catalog No.:BCC7120
CAS No.:264622-53-9
- MRS 1754
Catalog No.:BCC7473
CAS No.:264622-58-4
- 26-Deoxyactein
Catalog No.:BCN8076
CAS No.:264624-38-6
- Humulone
Catalog No.:BCN2682
CAS No.:26472-41-3
- Catharanthine Tartrate
Catalog No.:BCN2462
CAS No.:2648-21-5
- 6-Deoxyisojacareubin
Catalog No.:BCN7723
CAS No.:26486-92-0
- Cyclo(D-Phe-L-Pro)
Catalog No.:BCN4011
CAS No.:26488-24-4
- Clovanediol
Catalog No.:BCN5143
CAS No.:2649-64-1
- Clovanediol diacetate
Catalog No.:BCN5144
CAS No.:2649-68-5
- Nandrolone laurate
Catalog No.:BCC9088
CAS No.:26490-31-3
- Z-Gln-OH
Catalog No.:BCC2783
CAS No.:2650-64-8
The Deubiquitinase Inhibitor PR-619 Sensitizes Normal Human Fibroblasts to Tumor Necrosis Factor-related Apoptosis-inducing Ligand (TRAIL)-mediated Cell Death.[Pubmed:26757822]
J Biol Chem. 2016 Mar 11;291(11):5960-70.
TNF-related apoptosis-inducing ligand (TRAIL) is a potential cancer therapy that selectively targets cancer cell death while non-malignant cells remain viable. Using a panel of normal human fibroblasts, we characterized molecular differences in human foreskin fibroblasts and WI-38 TRAIL-resistant cells and marginally sensitive MRC-5 cells compared with TRAIL-sensitive human lung and colon cancer cells. We identified decreased caspase-8 protein expression and protein stability in normal fibroblasts compared with cancer cells. Additionally, normal fibroblasts had incomplete TRAIL-induced caspase-8 activation compared with cancer cells. We found that normal fibroblasts lack the ubiquitin modification of caspase-8 required for complete caspase-8 activation. Treatment with the deubiquitinase inhibitor PR-619 increased caspase-8 ubiquitination and caspase-8 enzymatic activity and sensitized normal fibroblasts to TRAIL-mediated apoptosis. Therefore, posttranslational regulation of caspase-8 confers resistance to TRAIL-induced cell death in normal cells through blockade of initiation of the extrinsic cell death pathway.
Inhibition of protein deubiquitination by PR-619 activates the autophagic pathway in OLN-t40 oligodendroglial cells.[Pubmed:23686611]
Cell Biochem Biophys. 2013 Sep;67(1):149-60.
Protein aggregate formation may be the result of an impairment of the protein quality control system, e.g., the ubiquitin proteasome system (UPS) and the lysosomal autophagic pathway. For proteasomal degradation, proteins need to be covalently modified by ubiquitin and deubiquitinated before the substrates are proteolytically degraded. Deubiquitination is performed by a large family of proteases, the deubiquitinating enzymes (DUBs). DUBs display a variety of functions and their inhibition may have pathological consequences. Using the broad specificity DUB inhibitor PR-619 we previously have shown that DUB inhibition leads to an overload of ubiquitinated proteins, to protein aggregate formation and subsequent inhibition of the UPS. This study was undertaken to investigate whether PR-619 modulates autophagic functions to possibly compensate the failure of the proteasomal system. Using the oligodendroglial cell line OLN-t40 and a new oligodendroglial cell line stably expressing GFP-LC3, we show that DUB inhibition leads to the activation of autophagy and to the recruitment of LC3 and of the ubiquitin binding protein p62 to the forming aggresomes without impairing the autophagic flux. Furthermore, PR-619 induced the transport of lysosomes to the forming aggregates in a process requiring an intact microtubule network. Further stimulation of autophagy by rapamycin did not prevent PR-619 aggregate formation but rather exerted cytotoxic effects. Hence, inhibition of DUBs by PR-619 activated the autophagic pathway supporting the hypothesis that the UPS and the autophagy-lysosomal pathway are closely linked together.
The small molecule inhibitor PR-619 of deubiquitinating enzymes affects the microtubule network and causes protein aggregate formation in neural cells: implications for neurodegenerative diseases.[Pubmed:22565157]
Biochim Biophys Acta. 2012 Nov;1823(11):2057-68.
A pathological hallmark of many neurodegenerative diseases is the aggregation of proteins. Protein aggregate formation may be linked to a failure of the ubiquitin proteasome system (UPS) and/or the autophagy pathway. The UPS involves the ubiquitination of proteins followed by proteasomal degradation. Deubiquitination of target proteins is performed by proteases called deubiquitinating proteins (DUBs). Inhibition of DUBs may lead to the dysregulation of homeostasis and have pathological consequences. To assess the effects of DUB-inhibition, we have used the oligodendroglial cell line, OLN-t40, stably expressing the longest human tau isoform. Cells were incubated with PR-619, a broad-range, reversible inhibitor of ubiquitin isopeptidases. Incubation with PR-619 led to morphological changes, the upregulation of heat shock proteins (HSP), including HSP70 and alphaB-crystallin, and to protein aggregates near the MTOC, containing ubiquitin, HSPs, and the ubiquitin binding protein p62, which may provide a link between the UPS and autophagy. Thus, inhibition of DUB activity caused stress responses and the formation of protein aggregates resembling pathological inclusions observed in aggregopathies. Furthermore, PR-619 led to the stabilization of the microtubule network, possibly through the modulation of tau phosphorylation, and small tau deposits assembled near the MTOC. Hence, organization and integrity of the cytoskeleton were affected, which is particularly important for the maintenance of the cellular architecture and intracellular transport processes, and essential for the functionality and survival of neural cells. Our data demonstrate that DUB inhibitors provide a useful tool to elucidate the manifold mechanisms of DUB functions in cells and their dysregulation in neurodegenerative diseases. This article is part of a Special Issue entitled: Ubiquitin Drug Discovery and Diagnostics.
Activity-based chemical proteomics accelerates inhibitor development for deubiquitylating enzymes.[Pubmed:22118674]
Chem Biol. 2011 Nov 23;18(11):1401-12.
Converting lead compounds into drug candidates is a crucial step in drug development, requiring early assessment of potency, selectivity, and off-target effects. We have utilized activity-based chemical proteomics to determine the potency and selectivity of deubiquitylating enzyme (DUB) inhibitors in cell culture models. Importantly, we characterized the small molecule PR-619 as a broad-range DUB inhibitor, and P22077 as a USP7 inhibitor with potential for further development as a chemotherapeutic agent in cancer therapy. A striking accumulation of polyubiquitylated proteins was observed after both selective and general inhibition of cellular DUB activity without direct impairment of proteasomal proteolysis. The repertoire of ubiquitylated substrates was analyzed by tandem mass spectrometry, identifying distinct subsets for general or specific inhibition of DUBs. This enabled identification of previously unknown functional links between USP7 and enzymes involved in DNA repair.


