N6-methyladenosine (m6A)adenylate cyclase regulator CAS# 1867-73-8 |
- SU14813
Catalog No.:BCC1971
CAS No.:627908-92-3
Quality Control & MSDS
3D structure
Package In Stock
Number of papers citing our products

| Cas No. | 1867-73-8 | SDF | Download SDF |
| PubChem ID | 1869 | Appearance | Powder |
| Formula | C11H15N5O4 | M.Wt | 281.27 |
| Type of Compound | N/A | Storage | Desiccate at -20°C |
| Synonyms | 6-Methyladenosine; N-Methyladenosine | ||
| Solubility | DMSO : ≥ 31 mg/mL (110.21 mM) *"≥" means soluble, but saturation unknown. | ||
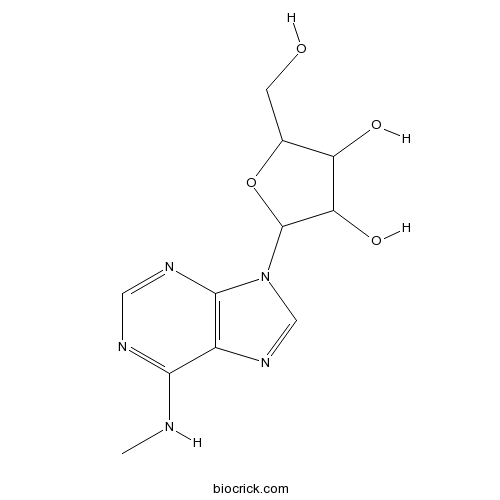
| Chemical Name | 2-(hydroxymethyl)-5-[6-(methylamino)purin-9-yl]oxolane-3,4-diol | ||
| SMILES | CNC1=NC=NC2=C1N=CN2C3C(C(C(O3)CO)O)O | ||
| Standard InChIKey | VQAYFKKCNSOZKM-UHFFFAOYSA-N | ||
| Standard InChI | InChI=1S/C11H15N5O4/c1-12-9-6-10(14-3-13-9)16(4-15-6)11-8(19)7(18)5(2-17)20-11/h3-5,7-8,11,17-19H,2H2,1H3,(H,12,13,14) | ||
| General tips | For obtaining a higher solubility , please warm the tube at 37 ℃ and shake it in the ultrasonic bath for a while.Stock solution can be stored below -20℃ for several months. We recommend that you prepare and use the solution on the same day. However, if the test schedule requires, the stock solutions can be prepared in advance, and the stock solution must be sealed and stored below -20℃. In general, the stock solution can be kept for several months. Before use, we recommend that you leave the vial at room temperature for at least an hour before opening it. |
||
| About Packaging | 1. The packaging of the product may be reversed during transportation, cause the high purity compounds to adhere to the neck or cap of the vial.Take the vail out of its packaging and shake gently until the compounds fall to the bottom of the vial. 2. For liquid products, please centrifuge at 500xg to gather the liquid to the bottom of the vial. 3. Try to avoid loss or contamination during the experiment. |
||
| Shipping Condition | Packaging according to customer requirements(5mg, 10mg, 20mg and more). Ship via FedEx, DHL, UPS, EMS or other couriers with RT, or blue ice upon request. | ||
| Description | N6-Methyladenosine is the most prevalent internal (non-cap) modification present in the messenger RNA (mRNA) of all higher eukaryotes.In Vitro:N6-methyladenosine (m6A) is selectively recognized by the human YTH domain family 2 (YTHDF2) protein to regulate mRNA degradation. N6-methyladenosine (m6A), a prevalent internal modification in the messenger RNA of all eukaryotes, is post-transcriptionally installed by m6A methyltransferase (e.g., MT-A70) within the consensus sequence of G(m6A)C (70%) or A(m6A)C (30%). N6-methyladenosine (m6A)-containing RNAs are greatly enriched in the YTHDF-bound portion and diminished in the flow-through portion[1]. N6-methyladenosine (m6A), the most abundant internal RNA modification, functions in diverse biological processes, including regulation of embryonic stem cell self-renewal and differentiation. N6-methyladenosine (m6A) is a large protein complex, consisting in part of methyltransferase-like 3 (METTL3) and methyltransferase-like 14 (METTL14) catalytic subunits[2]. References: | |||||

N6-methyladenosine (m6A) Dilution Calculator

N6-methyladenosine (m6A) Molarity Calculator
| 1 mg | 5 mg | 10 mg | 20 mg | 25 mg | |
| 1 mM | 3.5553 mL | 17.7765 mL | 35.553 mL | 71.1061 mL | 88.8826 mL |
| 5 mM | 0.7111 mL | 3.5553 mL | 7.1106 mL | 14.2212 mL | 17.7765 mL |
| 10 mM | 0.3555 mL | 1.7777 mL | 3.5553 mL | 7.1106 mL | 8.8883 mL |
| 50 mM | 0.0711 mL | 0.3555 mL | 0.7111 mL | 1.4221 mL | 1.7777 mL |
| 100 mM | 0.0356 mL | 0.1778 mL | 0.3555 mL | 0.7111 mL | 0.8888 mL |
| * Note: If you are in the process of experiment, it's necessary to make the dilution ratios of the samples. The dilution data above is only for reference. Normally, it's can get a better solubility within lower of Concentrations. | |||||

Calcutta University

University of Minnesota

University of Maryland School of Medicine

University of Illinois at Chicago

The Ohio State University

University of Zurich

Harvard University

Colorado State University

Auburn University

Yale University

Worcester Polytechnic Institute

Washington State University

Stanford University

University of Leipzig

Universidade da Beira Interior

The Institute of Cancer Research

Heidelberg University

University of Amsterdam

University of Auckland

TsingHua University

The University of Michigan

Miami University

DRURY University

Jilin University

Fudan University

Wuhan University

Sun Yat-sen University

Universite de Paris

Deemed University

Auckland University

The University of Tokyo

Korea University
N6-methyladenosine (m6A or N6m), an adenosine agonist, of an ED50 value of 17.250 µM [1], is a regulator of adenylate cyclase [2]. As a modification in the messenger RNA (mRNA), it is the most prevalent internal (non-cap) modification of mRNA in all higher eukaryotes. It is involved in the regulation of mRNA stability [3].
Adenylate cyclase catalyzes the formation of cyclic adenosine 3’,5’-monophosphate (cAMP) from ATP [4]. In Calu-6 cells, cAMP increased the stability of human rennin mRNA [5].
Concentration-dependently, N6m prevented the accumulation of hemoglobin and inhibited the initiation of commitment to terminal maturation. Treatment with N6m made cell growth slower, but it did not substantially inhibit DNA synthesis and markedly decrease viability and clonogenic potential of MEL cells. In MEL cells, treatment with N6m resulted in a decrease in the accumulation of βmajor globin mRNA in cytoplasm and affected the structural integrity of this mRNA. The inhibition of commitment induced by N6m was potentiated by adenine, l-homocysteine and/or l-methionine. Adenine, l-homocysteine and l-methionine are involved in the active methylation cycle [6].
In mammals, RNA m6A modification preferentially occurs within the consensus sequence RRACH (R = G or A; H = A, C or U) in 3’ UTRs and gene coding regions. That means m6A plays fundamental roles in the control of RNA processing and translational [7].
References:
[1]. Ribeiro JA and Sebastio AM. On the type of receptor involved in the inhibitory action of adenosine at the neuromuscular junction. Br J Pharmacol, 1985, 84(4):911-8.
[2]. Londos C, Wolff J and Cooper DMF. Adenosine as a regulator of adenylate cyclase[M]//Purinergic receptors. Springer Netherlands, 1981:287-323.
[3]. Wang X, Lu Z, Gomez A, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature, 2014, 505(7481): 117-120.
[4]. Salomon Y, Londos C and Rodbell M. A highly sensitive adenylate cyclase assay. Analytical biochemistry, 1974, 58(2): 541-548.
[5]. Sinn P L and Sigmund CD. Human renin mRNA stability is increased in response to cAMP in Calu-6 cells. Hypertension, 1999, 33(3): 900-905.
[6]. Vizirianakis IS, Wong W and Tsiftsoglou AS. Analysis of the inhibition of commitment of murine erythroleukemia (MEL) cells to terminal maturation by N 6-methyladenosine. Biochemical pharmacology, 1992, 44(5): 927-936.
[7]. Zhao X, Yang Y, Sun BF, et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell research, 2014, 24(12): 1403-1419.
- Ketamine hydrochloride
Catalog No.:BCC5982
CAS No.:1867-66-9
- H-D-Tyr(tBu)-OH
Catalog No.:BCC3137
CAS No.:186698-58-8
- Roscovitine (Seliciclib,CYC202)
Catalog No.:BCC1105
CAS No.:186692-46-6
- 2-NBDG
Catalog No.:BCC6530
CAS No.:186689-07-6
- 4-Methylcinnamic acid
Catalog No.:BCN5034
CAS No.:1866-39-3
- Allyl cinnamate
Catalog No.:BCC8812
CAS No.:1866-31-5
- 1,2-Bis(3-indenyl)ethane
Catalog No.:BCC8413
CAS No.:18657-57-3
- LY 344864
Catalog No.:BCC1716
CAS No.:186544-26-3
- Zibotentan (ZD4054)
Catalog No.:BCC2524
CAS No.:186497-07-4
- Alisol B
Catalog No.:BCN3364
CAS No.:18649-93-9
- Actein
Catalog No.:BCN1159
CAS No.:18642-44-9
- Psoralidin
Catalog No.:BCN5414
CAS No.:18642-23-4
- Alisol A 24-acetate
Catalog No.:BCN2344
CAS No.:18674-16-3
- Ginsenoside Rg5
Catalog No.:BCN3551
CAS No.:186763-78-0
- ML 10302 hydrochloride
Catalog No.:BCC7695
CAS No.:186826-17-5
- Moxifloxacin HCl
Catalog No.:BCC2507
CAS No.:186826-86-8
- 2B-(SP)
Catalog No.:BCC5817
CAS No.:186901-17-7
- Pafuramidine
Catalog No.:BCC1832
CAS No.:186953-56-0
- Sinapine
Catalog No.:BCN1815
CAS No.:18696-26-9
- N,N'-Bis(2-hydroxyethyl)oxamide
Catalog No.:BCC9061
CAS No.:1871-89-2
- NS309
Catalog No.:BCC1809
CAS No.:18711-16-5
- Clauszoline M
Catalog No.:BCN4683
CAS No.:187110-72-1
- Luliconazole
Catalog No.:BCC1711
CAS No.:187164-19-8
- Cyanidin-3-O-rutinoside chloride
Catalog No.:BCN3114
CAS No.:18719-76-1
SRAMP: prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features.[Pubmed:26896799]
Nucleic Acids Res. 2016 Jun 2;44(10):e91.
N(6)-methyladenosine (m(6)A) is a prevalent RNA methylation modification involved in the regulation of degradation, subcellular localization, splicing and local conformation changes of RNA transcripts. High-throughput experiments have demonstrated that only a small fraction of the m(6)A consensus motifs in mammalian transcriptomes are modified. Therefore, accurate identification of RNA m(6)A sites becomes emergently important. For the above purpose, here a computational predictor of mammalian m(6)A site named SRAMP is established. To depict the sequence context around m(6)A sites, SRAMP combines three random forest classifiers that exploit the positional nucleotide sequence pattern, the K-nearest neighbor information and the position-independent nucleotide pair spectrum features, respectively. SRAMP uses either genomic sequences or cDNA sequences as its input. With either kind of input sequence, SRAMP achieves competitive performance in both cross-validation tests and rigorous independent benchmarking tests. Analyses of the informative features and overrepresented rules extracted from the random forest classifiers demonstrate that nucleotide usage preferences at the distal positions, in addition to those at the proximal positions, contribute to the classification. As a public prediction server, SRAMP is freely available at http://www.cuilab.cn/sramp/.
Genome-Wide Location Analyses of N6-Methyladenosine Modifications (m(6)A-Seq).[Pubmed:28349453]
Methods Mol Biol. 2017;1562:45-53.
N(6)-methyladenosine-sequencing (m(6)A-seq) is a critical tool to obtain an unbiased genome-wide picture of m(6)A sites of modification at high resolution. It allows the study of the impact of various perturbations on m(6)A modification distribution and the study of m(6)A functions. Herein, we describe the m(6)A-seq protocol, which entails RNA immunoprecipitation (RIP) performed on fragmented poly(A) RNA utilizing anti-m(6)A antibodies. The captured/enriched m(6)A positive RNA fragments are subsequently sequenced by RNA-seq in parallel with background control non-immunoprecipitated input RNA fragments. Analyses reveal peaks of m(6)A enrichment containing sites of modifications analogous to chromatin modification immunoprecipitation experiments.


