AlrestatinAldose reductase inhibitor CAS# 51411-04-2 |
- Bimatoprost
Catalog No.:BCC4948
CAS No.:155206-00-1
- Misoprostol
Catalog No.:BCC5240
CAS No.:59122-46-2
Quality Control & MSDS
3D structure
Package In Stock
Number of papers citing our products

| Cas No. | 51411-04-2 | SDF | Download SDF |
| PubChem ID | 2120 | Appearance | Powder |
| Formula | C14H9NO4 | M.Wt | 255.23 |
| Type of Compound | N/A | Storage | Desiccate at -20°C |
| Synonyms | AY-22284 | ||
| Solubility | DMSO : 50 mg/mL (195.90 mM; Need ultrasonic) H2O : < 0.1 mg/mL (insoluble) | ||
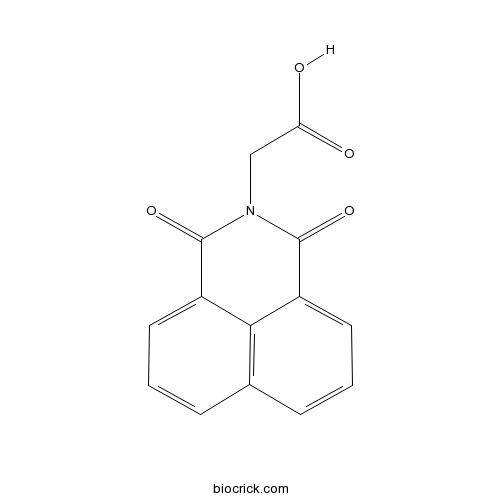
| Chemical Name | 2-(1,3-dioxobenzo[de]isoquinolin-2-yl)acetic acid | ||
| SMILES | C1=CC2=C3C(=C1)C(=O)N(C(=O)C3=CC=C2)CC(=O)O | ||
| Standard InChIKey | GCUCIFQCGJIRNT-UHFFFAOYSA-N | ||
| Standard InChI | InChI=1S/C14H9NO4/c16-11(17)7-15-13(18)9-5-1-3-8-4-2-6-10(12(8)9)14(15)19/h1-6H,7H2,(H,16,17) | ||
| General tips | For obtaining a higher solubility , please warm the tube at 37 ℃ and shake it in the ultrasonic bath for a while.Stock solution can be stored below -20℃ for several months. We recommend that you prepare and use the solution on the same day. However, if the test schedule requires, the stock solutions can be prepared in advance, and the stock solution must be sealed and stored below -20℃. In general, the stock solution can be kept for several months. Before use, we recommend that you leave the vial at room temperature for at least an hour before opening it. |
||
| About Packaging | 1. The packaging of the product may be reversed during transportation, cause the high purity compounds to adhere to the neck or cap of the vial.Take the vail out of its packaging and shake gently until the compounds fall to the bottom of the vial. 2. For liquid products, please centrifuge at 500xg to gather the liquid to the bottom of the vial. 3. Try to avoid loss or contamination during the experiment. |
||
| Shipping Condition | Packaging according to customer requirements(5mg, 10mg, 20mg and more). Ship via FedEx, DHL, UPS, EMS or other couriers with RT, or blue ice upon request. | ||
| Description | Specific inhibitor of aldose reductase (IC50 = 148 μM). Attenuates glucose-induced angiotensin II production in rat vascular smooth muscle in vitro. |

Alrestatin Dilution Calculator

Alrestatin Molarity Calculator
| 1 mg | 5 mg | 10 mg | 20 mg | 25 mg | |
| 1 mM | 3.918 mL | 19.5902 mL | 39.1803 mL | 78.3607 mL | 97.9509 mL |
| 5 mM | 0.7836 mL | 3.918 mL | 7.8361 mL | 15.6721 mL | 19.5902 mL |
| 10 mM | 0.3918 mL | 1.959 mL | 3.918 mL | 7.8361 mL | 9.7951 mL |
| 50 mM | 0.0784 mL | 0.3918 mL | 0.7836 mL | 1.5672 mL | 1.959 mL |
| 100 mM | 0.0392 mL | 0.1959 mL | 0.3918 mL | 0.7836 mL | 0.9795 mL |
| * Note: If you are in the process of experiment, it's necessary to make the dilution ratios of the samples. The dilution data above is only for reference. Normally, it's can get a better solubility within lower of Concentrations. | |||||

Calcutta University

University of Minnesota

University of Maryland School of Medicine

University of Illinois at Chicago

The Ohio State University

University of Zurich

Harvard University

Colorado State University

Auburn University

Yale University

Worcester Polytechnic Institute

Washington State University

Stanford University

University of Leipzig

Universidade da Beira Interior

The Institute of Cancer Research

Heidelberg University

University of Amsterdam

University of Auckland

TsingHua University

The University of Michigan

Miami University

DRURY University

Jilin University

Fudan University

Wuhan University

Sun Yat-sen University

Universite de Paris

Deemed University

Auckland University

The University of Tokyo

Korea University
Alrestatin is an inhibitor of aldose reductase, an enzyme involved in the pathogenesis of complications of diabetes mellitus, including diabetic neuropathy.
- Canthaxanthin
Catalog No.:BCC8139
CAS No.:514-78-3
- Biperiden
Catalog No.:BCC4274
CAS No.:514-65-8
- Ferruginol
Catalog No.:BCN3155
CAS No.:514-62-5
- Euphol
Catalog No.:BCN7790
CAS No.:514-47-6
- Tirucallol
Catalog No.:BCN7787
CAS No.:514-46-5
- Parkeol
Catalog No.:BCN3728
CAS No.:514-45-4
- Periplogenin
Catalog No.:BCN2656
CAS No.:514-39-6
- Abietic acid
Catalog No.:BCN2728
CAS No.:514-10-3
- Taraxerone
Catalog No.:BCN5636
CAS No.:514-07-8
- 2-(2-Aminoethyl)-1-methylpyrrolidine
Catalog No.:BCC8477
CAS No.:51387-90-7
- Boc-Methioninol
Catalog No.:BCC2720
CAS No.:51372-93-1
- Tris DBA
Catalog No.:BCC7685
CAS No.:51364-51-3
- Chikusetsusaponin IVa
Catalog No.:BCN3432
CAS No.:51415-02-2
- Odonicin
Catalog No.:BCN5637
CAS No.:51419-51-3
- 3-Methyladenine
Catalog No.:BCC3714
CAS No.:5142-23-4
- COG 133
Catalog No.:BCC1047
CAS No.:514200-66-9
- Cyclo(Tyr-Phe)
Catalog No.:BCN2423
CAS No.:5147-17-1
- Deoxynivalenol
Catalog No.:BCC7832
CAS No.:51481-10-8
- Cimetidine
Catalog No.:BCC4527
CAS No.:51481-61-9
- PMX 205
Catalog No.:BCC8039
CAS No.:514814-49-4
- H-Tyr(tBu)-OMe.HCl
Catalog No.:BCC2672
CAS No.:51482-39-4
- Sclareol
Catalog No.:BCN2395
CAS No.:515-03-7
- (+)-Turicine
Catalog No.:BCC8361
CAS No.:515-24-2
- Cochinchinenin A
Catalog No.:BCN3496
CAS No.:221696-69-1
Inhibition of human brain aldose reductase and hexonate dehydrogenase by alrestatin and sorbinil.[Pubmed:6808090]
J Neurochem. 1982 Sep;39(3):810-4.
Human brain aldose reductase and hexonate dehydrogenase are inhibited by Alrestatin (AY 22,284) and sorbinil (CP 45,634). Inhibition by Alrestatin is noncompetitive for both enzymes, and slightly stronger for hexonate dehydrogenase (KI values 52-250 microM) than for aldose reductase (KI values 170-320 microM). Sorbinil inhibits hexonate dehydrogenase far more potently than aldose reductase, KI values being 5 5 microM for hexonate dehydrogenase and 150 microM for aldose reductase. The inhibition of hexonate dehydrogenase by sorbinil is noncompetitive with respect to both aldehyde and NADPH substrates, and is thus kinetically similar to the inhibition by Alrestatin. However, sorbinil inhibition of aldose reductase is uncompetitive with respect to glyceraldehyde and noncompetitive with NADPH as the varied substrate. Inhibition of human brain aldose reductase by these two inhibitors is much less potent than that reported for the enzyme from other sources.
The alrestatin double-decker: binding of two inhibitor molecules to human aldose reductase reveals a new specificity determinant.[Pubmed:9405046]
Biochemistry. 1997 Dec 23;36(51):16134-40.
It is generally expected that only one inhibitor molecule will bind to an enzyme active site. In fact, specific drug design theories depend upon this assumption. Here, we report the binding of two molecules of an inhibitor to the same active site which we observed in the 1.8 A resolution structure of the drug Alrestatin bound to a mutant of human aldose reductase. The two molecules of Alrestatin bind to the active site in a stacked arrangement (a double-decker). This stack positions the carboxylic acid of one drug molecule near the NADP+ cofactor at a previously determined anion binding site and the carboxylic acid of the second drug molecule near the carboxy-terminal tail of the enzyme. We propose that interactions of inhibitors with the carboxy-terminal loop of aldose reductase are critical for the development of inhibitors that are able to discriminate between aldose reductase and other members of the aldo-keto reductase superfamily. This finding suggests a new direction for the introduction of specificity to aldose reductase-targeted drugs.
Human placenta aldose reductase. Forms sensitive and insensitive to inhibition by alrestatin.[Pubmed:6427599]
Mol Pharmacol. 1984 May;25(3):425-30.
The inhibition of aldose reductase from a human source by Alrestatin was studied. The enzyme from placenta was purified to apparent homogeneity by (NH4)2SO4 precipitation, DEAE-cellulose chromatography, electrofocusing, and affinity chromatography. This enzyme from human or rat placenta at the (NH4)2SO4 state of purification was relatively insensitive to Alrestatin (IC50 greater than 50 microM). On purification by electrofocusing, however, human or rat placenta aldose reductase exhibited a marked increase in its sensitivity to Alrestatin (IC50 = 1.0 microM). In contrast to human or rat placenta aldose reductase, rat lens aldose reductase was equally sensitive to Alrestatin at the corresponding stages of purification (IC50 = 1.0 microM). Experiments in which the sensitive and insensitive forms of placenta aldose reductase were mixed revealed that the difference in susceptibility to Alrestatin could not be attributed to nonspecific binding of Alrestatin by proteins present in the (NH4)2SO4 fraction. A heat-inactivated (NH4)2SO4 fraction of human placenta aldose reductase added to the sensitive placenta enzyme from human or rats caused a time-dependent conversion to the insensitive form of aldose reductase. This suggested that a heat-stable dissociable factor, associated with placenta aldose reductase at the crude stage, may be responsible for the insensitivity to Alrestatin. This insensitivity could be of pharmacological significance if it is relevant in vivo and it exists in tissues where aldose reductase plays a physiological role.
Mechanism of aldose reductase inhibition: binding of NADP+/NADPH and alrestatin-like inhibitors.[Pubmed:8003482]
Biochemistry. 1994 Jun 14;33(23):7157-65.
Aldose reductase enfolds NADP+/NADPH via a complex loop mechanism, with cofactor exchange being the rate-limiting step for the overall reaction. This study measures the binding constants of these cofactors in the wild-type enzyme, as well as a variety of active-site mutants (C298A, Y48H, Y48F, Y209F, H110A, W219A, and W20A), and seeks to identify the binding site and mechanism of the aldose reductase inhibitor Alrestatin in the recombinant human enzyme. All the mutant enzymes, regardless of their enzyme activities, have normal or only slightly elevated coenzyme binding constants, suggesting a tertiary structure similar to that of the wild-type enzyme. Binding of Alrestatin was detected by fluorescence assays, and by an ultrafiltration assay which measures the fraction of unbound Alrestatin. Alrestatin binds preferentially to the enzyme/NADP+ complex, consistent with the steady-state inhibition pattern. Alrestatin binding and enzyme inhibition were abolished in the Tyr48 mutants Y48F and Y48H, implicating the positively charged anion well formed by the Asp43-/Lys77+/Tyr48(0)/NADP+ complex in inhibitor binding (Harrison et al., 1994; Bohren et al., 1994). The enzyme mutant W20A severely affected the inhibitory potencies of a variety of commercially developed aldose reductase inhibitors (zopolrestat, tolrestat, FK366, AL1576, Alrestatin, ponalrestat, and sorbinil). Inhibition by citrate, previously shown to bind to the positively charged anion well, was not affected by this mutation. Inhibitors with flexible double aromatic ring systems (Zopolrestat, FK366, and ponalrestat) were less affected than others possessing a single aromatic ring system, suggesting that the additional pharmacophor ring system stabilizes the inhibitor by interaction at some other hydrophobic site.(ABSTRACT TRUNCATED AT 250 WORDS)
Mechanism of high glucose induced angiotensin II production in rat vascular smooth muscle cells.[Pubmed:17626897]
Circ Res. 2007 Aug 31;101(5):455-64.
Angiotensin II (Ang II), a circulating hormone that can be synthesized locally in the vasculature, has been implicated in diabetes-associated vascular complications. This study was conducted to determine whether high glucose (HG) (approximately 23.1 mmol/L), a diabetic-like condition, stimulates Ang II generation and the underlying mechanism of its production in rat vascular smooth muscle cells. The contribution of various enzymes involved in Ang II generation was investigated by silencing their expression with small interfering RNA in cells exposed to normal glucose (approximately 4.1 mmol/L) and HG. Angiotensin I (Ang I) was generated from angiotensinogen by cathepsin D in the presence of normal glucose or HG. Although HG did not affect the rate of angiotensinogen conversion, it decreased expression of angiotensin-converting enzyme (ACE), downregulated ACE-dependent Ang II generation, and upregulated rat vascular chymase-dependent Ang II generation. The ACE inhibitor captopril reduced Ang II levels in the media by 90% in the presence of normal glucose and 19% in HG, whereas rat vascular chymase silencing reduced Ang II production in cells exposed to HG but not normal glucose. The glucose transporter inhibitor cytochalasin B, the aldose reductase inhibitor Alrestatin, and the advanced glycation end product formation inhibitor aminoguanidine attenuated HG-induced Ang II generation. HG caused a transient increase in extracellular signal-regulated kinase (ERK)1/2 phosphorylation, and ERK1/2 inhibitors reduced Ang II accumulation by HG. These data suggest that polyol pathway metabolites and AGE can stimulate rat vascular chymase activity via ERK1/2 activation and increase Ang II production. In addition, decreased Ang II degradation, which, in part, could be attributable to a decrease in angiotensin-converting enzyme 2 expression observed in HG, contributes to increased accumulation of Ang II in vascular smooth muscle cells by HG.
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity.[Pubmed:8916913]
Biochemistry. 1996 Nov 12;35(45):14276-80.
Human aldehyde reductase has a preference for carboxyl group-containing negatively charged substrates. It belongs to the NADPH-dependent aldo-keto reductase superfamily whose members are in part distinguished by unique C-terminal loops. To probe the role of the C-terminal loops in determining substrate specificities in these enzymes, two arginine residues, Arg308 and Arg311, located in the C-terminal loop of aldehyde reductase, and not found in any other C-terminal loop, were replaced with alanine residues. The catalytic efficiency of the R311A mutant for aldehydes containing a carboxyl group is reduced 150-250-fold in comparison to that of the wild-type enzyme, while substrates not containing a negative charge are unaffected. The R311A mutant is also significantly less sensitive to inhibition by dicarboxylic acids, indicating that Arg311 interacts with one of the carboxyl groups. The inhibition pattern indicates that the other carboxyl group binds to the anion binding site formed by Tyr49, His112, and the nicotinamide moiety of NADP+. The correlation between inhibitor potency and the length of the dicarboxylic acid molecules suggests a distance of approximately 10 A between the amino group of Arg311 and the anion binding site in the aldehyde reductase molecule. The sensitivity of inhibition of the R311A mutant by several commercially available aldose reductase inhibitors (ARIs) was variable, with tolrestat and zopolrestat becoming more potent inhibitors (30- and 5-fold, respectively), while others remained the same or became less potent. The catalytic properties, substrate specificity, and susceptibility to inhibition of the R308A mutant remained similar to that of the wild-type enzyme. The data provide direct evidence for C-terminal loop participation in determining substrate and inhibitor specificity of aldo-keto reductases and specifically identifies Arg311 as the basis for the carboxyl-containing substrate preference of aldehyde reductase.


