Glycine max
Glycine max
1. The products in our compound library are selected from thousands of unique natural products; 2. It has the characteristics of diverse structure, diverse sources and wide coverage of activities; 3. Provide information on the activity of products from major journals, patents and research reports around the world, providing theoretical direction and research basis for further research and screening; 4. Free combination according to the type, source, target and disease of natural product; 5. The compound powder is placed in a covered tube and then discharged into a 10 x 10 cryostat; 6. Transport in ice pack or dry ice pack. Please store it at -20 °C as soon as possible after receiving the product, and use it as soon as possible after opening.
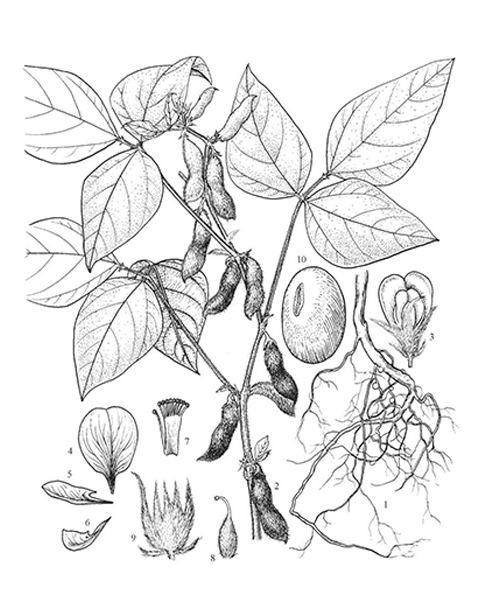
Natural products/compounds from Glycine max
- Cat.No. Product Name CAS Number COA
-
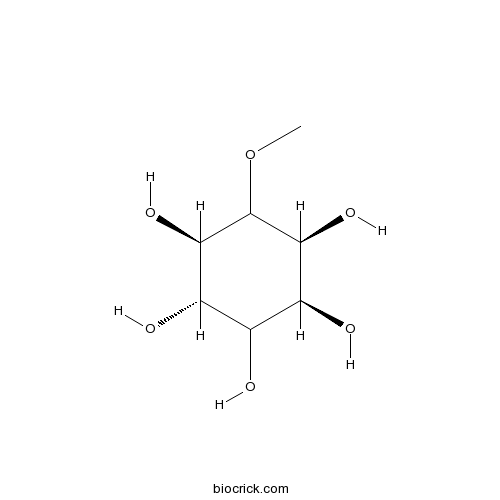
BCN5842
D-Pinitol10284-63-6
Instructions
-
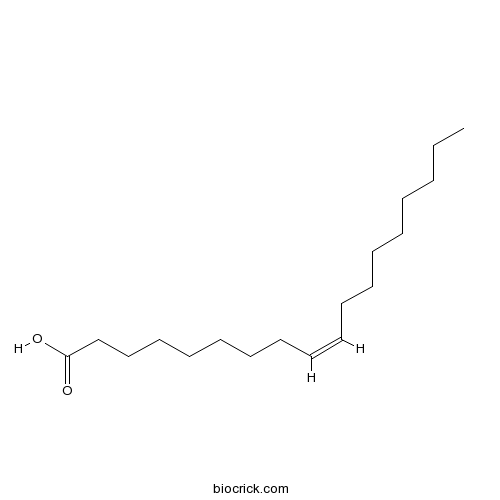
BCN7159
Oleic acid112-80-1
Instructions
-
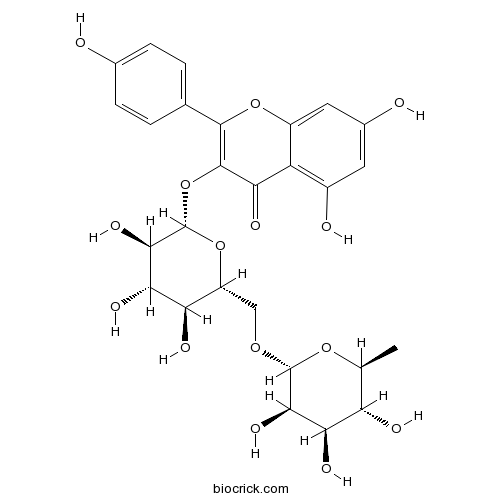
BCN1126
Nicotiflorin17650-84-9
Instructions
-
BCN2910
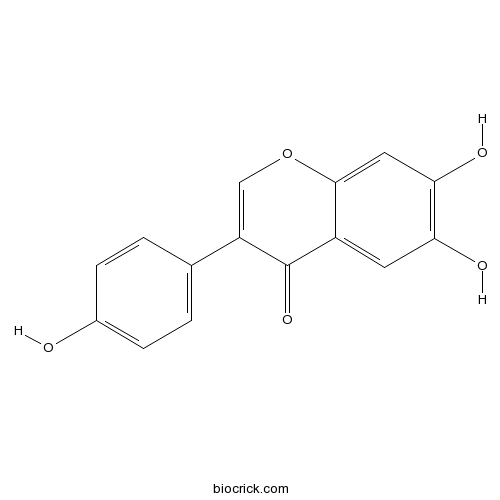
6,7,4'-Trihydroxyisoflavone17817-31-1
Instructions
-
BCN5423
Vitexin3681-93-4
Instructions

-
BCN5895
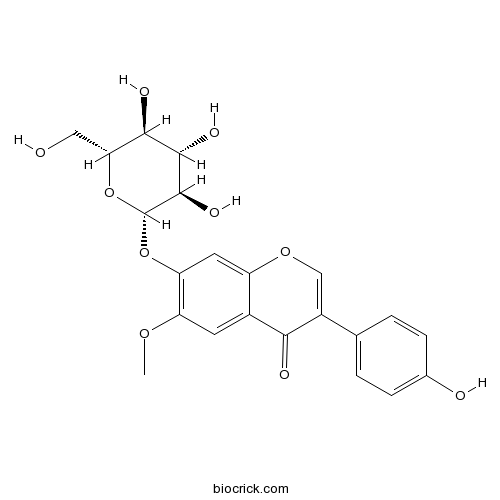
Glycitin40246-10-4
Instructions
-
BCN5896
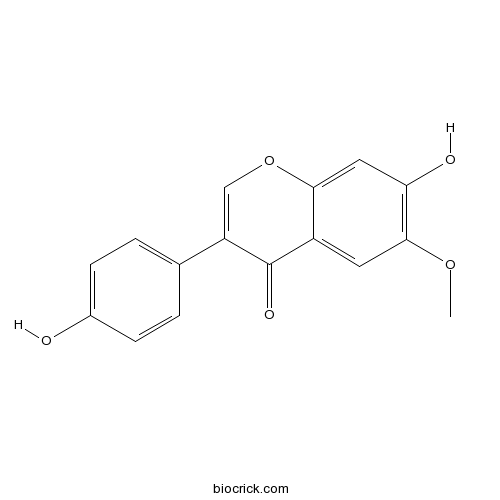
Glycitein40957-83-3
Instructions
-
BCN5590
Daidzein486-66-8
Instructions

-
BCN2772
6''-O-Malonylgenistin51011-05-3
Instructions

-
BCN2598
Soyasaponin Bb51330-27-9
Instructions

-
BCN5658
Apigenin520-36-5
Instructions

-
BCN1231
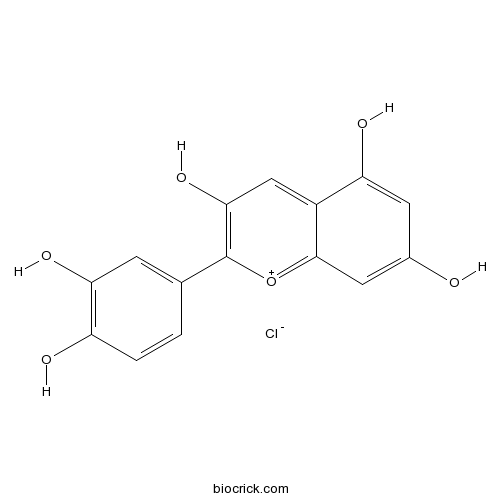
Cyanidin Chloride528-58-5
Instructions
-
BCN2396
Genistin529-59-9
Instructions

-
BCN5891
Daidzin552-66-9
Instructions

-
BCN8328
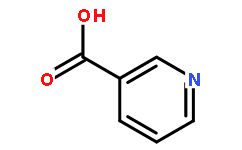
Nicotinic acid59-67-6
Instructions
-
BCN3818
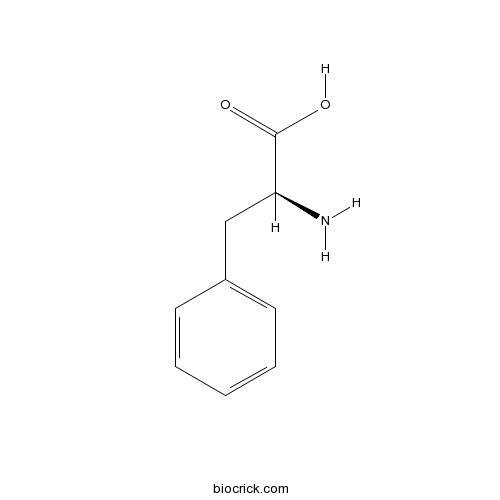
L-Phenylalanine63-91-2
Instructions
-
BCN1230
Cyanidin-3-O-glucoside chloride7084-24-4
Instructions

-
BCC3111
H-Trp-OH73-22-3
Instructions

-
BCN1015
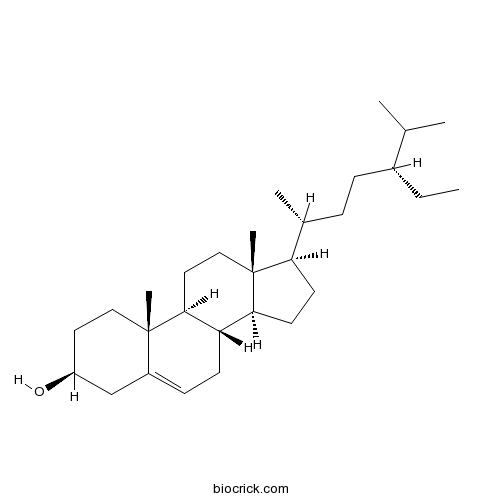
Beta-Sitosterol83-46-5
Instructions
GmBTB/POZ, a novel BTB/POZ domain-containing nuclear protein, positively regulates the response of soybean to Phytophthora sojae infection.[Pubmed: 30113770]
Phytophthora sojae is a destructive pathogen of soybean [Glycine max (L.) Merr.] causing stem and root rot to soybean plants worldwide. However, the pathogenesis and molecular mechanism of plant defense responses against P. sojae are largely unclear. Herein, we document the underlying mechanisms and function of a novel BTB/POZ protein in soybean, GmBTB/POZ, which contains a BTB/POZ domain found in certain animal transcriptional regulators, in host soybean plants in response to P. sojae. It is located in the cell nucleus and transcriptionally up-regulated by P. sojae. Overexpression of GmBTB/POZ in soybean resulted in enhanced resistance to P. sojae. The activities and expression levels of enzymatic SOD and POD antioxidants were significantly higher in GmBTB/POZ-OE transgenic soybean plants than in wild-type (WT) plants treated with sterile water or infected with P. sojae. The transcript levels of defense-associated genes were also higher in the overexpressed plants than in WT upon infection. Moreover, salicylic acid (SA) levels and the transcript levels of SA biosynthesis-related genes were markedly higher in GmBTB/POZ-OE transgenic soybean than in WT, but there were almost no differences in jasmonic acid (JA) levels or JA biosynthesis-related gene expression among GmBTB/POZ-OE and WT soybean lines. Furthermore, exogenous SA application induced the expression of GmBTB/POZ and inhibited the increase in P. sojae biomass in both WT and GmBTB/POZ-OE transgenic soybean plants. Taken together, these results suggest that GmBTB/POZ plays a positive role in P. sojae resistance and the defense response in soybean via a process that might be dependent on SA. This article is protected by copyright. All rights reserved.
Transcriptomic comparison reveals genetic variation potentially underlying seed developmental evolution of soybeans.[Pubmed: 30113693]
Soybean (Glycine max) was domesticated from its wild relative Glycine soja. However, the genetic variations underlying soybean domestication are not well known. Comparative transcriptomics revealed that a small portion of the orthologous genes might have been fast-evolving. In contrast, three gene expression clusters were identified as divergent by their expression patterns, which occupied 37.44% of the total genes, hinting at an essential role for gene expression alteration in soybean domestication. Moreover, the most divergent stage in gene expression between wild and cultivated soybeans occurred during seed development around the cotyledon stage (15 days after fertilization, G15). A module in which the coexpressed genes were significantly downregulated at G15 of wild soybeans was identified. The divergent clusters and modules included substantial differentially expressed genes (DEGs) between wild and cultivated soybeans related to cell division, storage compound accumulation, hormone-response, and seed maturation processes. Chromosomal-linked DEGs, quantitative trait loci controlling seed weight and oil content, and selection sweeps revealed candidate DEGs at G15 in the fruit-related divergence of G. max and G. soja. Our work establishes a transcriptomic selection mechanism for altering gene expression during soybean domestication, thus shedding light on the molecular networks underlying soybean seed development and breeding strategy.
In silico characterization and expression profiling of the diacylglycerol acyltransferase gene family (DGAT1, DGAT2, DGAT3 and WS/DGAT) from oil palm, Elaeis guineensis.[Pubmed: 30107884]
The diacylglycerol acyltransferases (DGAT) (diacylglycerol:acyl-CoA acyltransferase, EC 2.3.1.20) are a key group of enzymes that catalyse the final and usually the most important rate-limiting step of triacylglycerol biosynthesis in plants and other organisms. Genes encoding four distinct functional families of DGAT enzymes have been characterised in the genome of the African oil palm, Elaeis guineensis. The contrasting features of the various isoforms within the four families of DGAT genes, namely DGAT1, DGAT2, DGAT3 and WS/DGAT are presented both in the oil palm itself and, for comparative purposes, in 12 other oil crop or model/related plants, namely Arabidopsis thaliana, Brachypodium distachyon, Brassica napus, Elaeis oleifera, Glycine max, Gossypium hirsutum, Helianthus annuus, Musa acuminata, Oryza sativa, Phoenix dactylifera, Sorghum bicolor, and Zea mays. The oil palm genome contains respectively three, two, two and two distinctly expressed functional copies of the DGAT1, DGAT2, DGAT3 and WS/DGAT genes. Phylogenetic analyses of the four DGAT families showed that the E. guineensis genes tend to cluster with sequences from P. dactylifera and M. acuminata rather than with other members of the Commelinid monocots group, such as the Poales which include the major cereal crops such as rice and maize. Comparison of the predicted DGAT protein sequences with other animal and plant DGATs was consistent with the E. guineensis DGAT1 being ER located with its active site facing the lumen while DGAT2, although also ER located, had a predicted cytosol-facing active site. In contrast, DGAT3 and some (but not all) WS/DGAT in E. guineensis are predicted to be soluble, cytosolic enzymes. Evaluation of E. guineensis DGAT gene expression in different tissues and developmental stages suggests that the four DGAT groups have distinctive physiological roles and are particularly prominent in developmental processes relating to reproduction, such as flowering, and in fruit/seed formation especially in the mesocarp and endosperm tissues.


