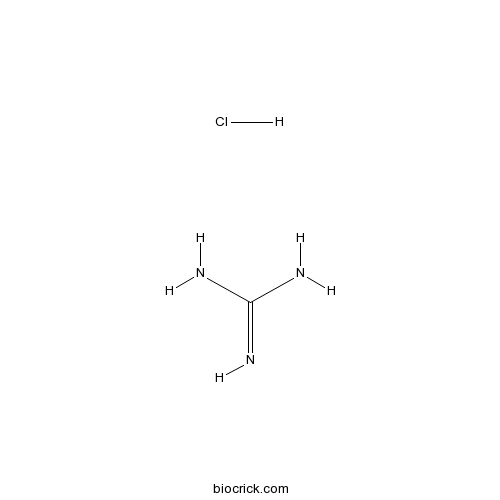
Guanidine HClCAS# 50-01-1 |
- Salmefamol
Catalog No.:BCC1919
CAS No.:18910-65-1
- (R,R)-Formoterol
Catalog No.:BCC1293
CAS No.:67346-49-0
- Doxazosin Mesylate
Catalog No.:BCC1257
CAS No.:77883-43-3
- Medetomidine
Catalog No.:BCC1736
CAS No.:86347-14-0
Quality Control & MSDS
3D structure
Package In Stock
Number of papers citing our products

| Cas No. | 50-01-1 | SDF | Download SDF |
| PubChem ID | 5742 | Appearance | Powder |
| Formula | CH6ClN3 | M.Wt | 95.53 |
| Type of Compound | N/A | Storage | Desiccate at -20°C |
| Synonyms | Guanidinium chloride; Aminoformamidine Hydrochloride | ||
| Solubility | DMSO : ≥ 45 mg/mL (113.36 mM) *"≥" means soluble, but saturation unknown. | ||
| Chemical Name | guanidine;hydrochloride | ||
| SMILES | [H+].[Cl-].NC(N)=N | ||
| Standard InChIKey | PJJJBBJSCAKJQF-UHFFFAOYSA-N | ||
| Standard InChI | InChI=1S/CH5N3.ClH/c2-1(3)4;/h(H5,2,3,4);1H | ||
| General tips | For obtaining a higher solubility , please warm the tube at 37 ℃ and shake it in the ultrasonic bath for a while.Stock solution can be stored below -20℃ for several months. We recommend that you prepare and use the solution on the same day. However, if the test schedule requires, the stock solutions can be prepared in advance, and the stock solution must be sealed and stored below -20℃. In general, the stock solution can be kept for several months. Before use, we recommend that you leave the vial at room temperature for at least an hour before opening it. |
||
| About Packaging | 1. The packaging of the product may be reversed during transportation, cause the high purity compounds to adhere to the neck or cap of the vial.Take the vail out of its packaging and shake gently until the compounds fall to the bottom of the vial. 2. For liquid products, please centrifuge at 500xg to gather the liquid to the bottom of the vial. 3. Try to avoid loss or contamination during the experiment. |
||
| Shipping Condition | Packaging according to customer requirements(5mg, 10mg, 20mg and more). Ship via FedEx, DHL, UPS, EMS or other couriers with RT, or blue ice upon request. | ||
| Description | Protein denaturant. |

Guanidine HCl Dilution Calculator

Guanidine HCl Molarity Calculator
| 1 mg | 5 mg | 10 mg | 20 mg | 25 mg | |
| 1 mM | 10.4679 mL | 52.3396 mL | 104.6792 mL | 209.3583 mL | 261.6979 mL |
| 5 mM | 2.0936 mL | 10.4679 mL | 20.9358 mL | 41.8717 mL | 52.3396 mL |
| 10 mM | 1.0468 mL | 5.234 mL | 10.4679 mL | 20.9358 mL | 26.1698 mL |
| 50 mM | 0.2094 mL | 1.0468 mL | 2.0936 mL | 4.1872 mL | 5.234 mL |
| 100 mM | 0.1047 mL | 0.5234 mL | 1.0468 mL | 2.0936 mL | 2.617 mL |
| * Note: If you are in the process of experiment, it's necessary to make the dilution ratios of the samples. The dilution data above is only for reference. Normally, it's can get a better solubility within lower of Concentrations. | |||||

Calcutta University

University of Minnesota

University of Maryland School of Medicine

University of Illinois at Chicago

The Ohio State University

University of Zurich

Harvard University

Colorado State University

Auburn University

Yale University

Worcester Polytechnic Institute

Washington State University

Stanford University

University of Leipzig

Universidade da Beira Interior

The Institute of Cancer Research

Heidelberg University

University of Amsterdam

University of Auckland

TsingHua University

The University of Michigan

Miami University

DRURY University

Jilin University

Fudan University

Wuhan University

Sun Yat-sen University

Universite de Paris

Deemed University

Auckland University

The University of Tokyo

Korea University
Guanidine HCl, the crystalline compound of strong alkalinity formed by the oxidation of guanine, is a normal product of protein metabolism and a protein denaturant.
- Tioxolone
Catalog No.:BCC2316
CAS No.:4991-65-5
- 2,4-Pyridinedicarboxylic Acid
Catalog No.:BCC6483
CAS No.:499-80-9
- 5-Isopropyl-2-methylphenol
Catalog No.:BCN2633
CAS No.:499-75-2
- beta-Thujaplicin
Catalog No.:BCN3895
CAS No.:499-44-5
- IsoMaltose
Catalog No.:BCN8321
CAS No.:499-40-1
- Corydamine
Catalog No.:BCN3366
CAS No.:49870-84-0
- Erythroskyrin
Catalog No.:BCN1836
CAS No.:4987-27-3
- EX 527 (SEN0014196)
Catalog No.:BCC2223
CAS No.:49843-98-3
- Tobramycin Sulfate
Catalog No.:BCC5633
CAS No.:49842-07-1
- Ethyl Nipecotate
Catalog No.:BCC3272
CAS No.:5006-62-2
- Scopine
Catalog No.:BCN1940
CAS No.:498-45-3
- 1,6-Anhydro-β-D-glucose
Catalog No.:BCC8427
CAS No.:498-07-7
- Dexamethasone (DHAP)
Catalog No.:BCC1184
CAS No.:50-02-2
- Cortisone acetate
Catalog No.:BCC4771
CAS No.:50-04-4
- Mitomycin C
Catalog No.:BCC2388
CAS No.:50-07-7
- Ergocalciferol
Catalog No.:BCN2208
CAS No.:50-14-6
- Cyclophosphamide
Catalog No.:BCC1185
CAS No.:50-18-0
- Corticosterone
Catalog No.:BCN2203
CAS No.:50-22-6
- Hydrocortisone
Catalog No.:BCN2192
CAS No.:50-23-7
- Prednisolone
Catalog No.:BCC4830
CAS No.:50-24-8
- Estriol
Catalog No.:BCN2235
CAS No.:50-27-1
- beta-Estradiol
Catalog No.:BCN2194
CAS No.:50-28-2
- Phenylbutazone
Catalog No.:BCC4822
CAS No.:50-33-9
- Thalidomide
Catalog No.:BCC2248
CAS No.:50-35-1
Guanidine-HCl dependent structural unfolding of M-crystallin: fluctuating native state like topologies and intermolecular association.[Pubmed:23284604]
PLoS One. 2012;7(12):e42948.
Numerous experimental techniques and computational studies, proposed in recent times, have revolutionized the understanding of protein-folding paradigm. The complete understanding of protein folding and intermediates are of medical relevance, as the aggregation of misfolding proteins underlies various diseases, including some neurodegenerative disorders. Here, we describe the unfolding of M-crystallin, a betagamma-crystallin homologue protein from archaea, from its native state to its denatured state using multidimensional NMR and other biophysical techniques. The protein, which was earlier characterized to be a predominantly beta-sheet protein in its native state, shows different structural propensities (alpha and beta), under different denaturing conditions. In 2 M GdmCl, the protein starts showing two distinct sets of peaks, with one arising from a partially unfolded state and the other from a completely folded state. The native secondary structural elements start disappearing as the denaturant concentration approaches 4 M. Subsequently, the protein is completely unfolded when the denaturant concentration is 6 M. The (15)N relaxation data (T(1)/T(2)), heteronuclear (1)H-(15)N Overhauser effects (nOes), NOESY data, and other biophysical data taken together indicate that the protein shows a consistent, gradual change in its structural and motional preferences with increasing GdmCl concentration.
Differing structural characteristics of molten globule intermediate of peanut lectin in urea and guanidine-HCl.[Pubmed:22595796]
Int J Biol Macromol. 2012 Oct;51(3):188-95.
The structural characteristics of exclusive equilibrium molten globule-like intermediate formed during peanut lectin unfolding in urea and guanidine hydrochloride (GdnHCl) have been investigated by size-exclusion chromatography, circular dichroism, fluorescence, phosphorescence, and chemical modification. The elution behavior and 8-anilino-1-naphthalenesulfonate binding indicate a less compact tertiary structure in urea than in GdnHCl. Further, the urea-induced intermediate reveals perturbed, nonnative typical beta-sheet conformation in contrast to native-like atypical beta-structure in GdnHCl. N-bromosuccinimide oxidation shows that none of three tryptophan residues is modified for GdnHCl-induced intermediate while one gets oxidized in urea. Such difference in tryptophan environment is supported by acrylamide quenching (Stern-Volmer constant being 3.2 and 5.8 M(-1) respectively), and phosphorescence studies at 77 K which show a blue-shift of (0, 0) band from 412.4 nm (GdnHCl) to 411.4 nm (urea). These results may provide important insight into the differential effects of GdnHCl and urea on the structural characteristics of intermediate state(s) in protein folding.
Single mother-daughter pair analysis to clarify the diffusion properties of yeast prion Sup35 in guanidine-HCl-treated [PSI] cells.[Pubmed:19674118]
Genes Cells. 2009 Sep;14(9):1045-54.
The yeast prion [PSI(+)] is a protein-based heritable element, in which aggregates of Sup35 protein are transmitted to daughter cells in a non-Mendelian manner. To elucidate the mechanism of the transmission, we have developed methods to directly analyse the dynamics of Sup35 fused with GFP in single mother-daughter pairs. As it is known that the treatment of yeast cells with guanidine hydrochloride (GuHCl) cures [PSI(+)] by perturbing Hsp104, a prion-remodelling factor, we analysed the diffusion profiles of Sup35-GFP in GuHCl-treated [PSI(+)] cells using fluorescence correlation spectroscopy (FCS). FCS analyses revealed that Sup35-GFP diffusion in the daughter cells was faster; that is, the Sup35-GFP particle was smaller, than that in the mother [PSI(+)] cells, and it eventually reached the diffusion profiles in [psi(-)] cells. We then analysed the flux of Sup35-GFP oligomers from mother to daughter [PSI(+)] cells in the presence of GuHCl, using a modified fluorescent recovery after photobleaching technique, and found that the flux of the diffuse oligomers was completely inhibited. The noninvasive methods described here can be applied to other protein-based transmissible systems inside living cells.
Dynamics of geminate rebinding of CO to cytochrome c in guanidine HCl probed by femtosecond vibrational spectroscopy.[Pubmed:23590118]
J Phys Chem B. 2013 May 2;117(17):4934-44.
Femtosecond vibrational spectroscopy was used to probe the rebinding dynamics of CO to cytochrome c (Cytc) in 1.8 and 7 M Guanidine HCl (GdnHCl) after photodeligation of the corresponding CO-bound protein in D2O buffer (pD = 7.4) at 283 K. Geminate rebinding (GR) dynamics of CO to the folded Cytc in 1.8 M GdnHCl (nCytc) is similar to that to chemically modified cytochrome c (cCytc), suggesting that the overall conformations of nCytcCO and cCytcCO are similar. About 86% of the dissociated CO molecules were geminately rebound to nCytc nonexponentially within 1 ns. The efficient GR of CO to the folded Cytc can be attributed to the organized protein matrix near the active site of nCytc that provides an efficient trap for the diffusing CO ligand after photodissociation. Although the concentration of nCytc did not affect its GR yield of CO, GR yield of CO to the unfolded Cytc in 7 M GdnHCl (uCytc) increased from 5 to 30% as the protein concentration increased from 0.3 to 9 mM. Time-resolved spectra of the (13)CO dissociated from both 9 mM nCytc(13)CO and 9 mM uCytc(13)CO showed a growing band with a peak at 2090 cm(-1) on the picosecond time scale, which was assigned to (13)CO in D2O solvent. At 1 ns, the fraction of the CO band in the solvent was about 10% of the nascent photodeligated protein in nCytc and more than 50% in the concentrated uCytc. Whereas a small opening in the active site of nCytc is responsible for the ultrafast escape of CO to solution in the folded protein, a large fraction of the CO escape to the solvent in uCytc results from the denatured structure of the active site in the unfolded protein. The spectrum of the CO dissociated from the concentrated uCytcCO contained a band that decayed as efficiently as that for the folded protein, suggesting that some fraction of uCytcCO might form aggregates even in 7 M denaturant, such that the aggregate acts as an efficient trap for the diffusing CO after deligation. No hint of precipitate in the concentrated uCytcCO and protein refolding upon dilution of the GdnHCl indicate that the aggregate does not grow continuously but remains as a soluble oligomer. The delayed appearance of the solvated CO and the inefficient GR of CO in uCytcCO suggest that the monomeric unfolded CytcCO so loosely arranged that the protein matrix cannot trap CO efficiently but the bound CO is still buried within hydrophobic residues even under the harsh denaturation condition.


