PLpro inhibitorCAS# 1093070-14-4 |
- Reparixin
Catalog No.:BCC1885
CAS No.:266359-83-5
- Reparixin L-lysine salt
Catalog No.:BCC1886
CAS No.:266359-93-7
- SCH 527123
Catalog No.:BCC1932
CAS No.:473727-83-2
- AMD-070
Catalog No.:BCC1357
CAS No.:558447-26-0
- AMD-070 hydrochloride
Catalog No.:BCC1358
CAS No.:880549-30-4
Quality Control & MSDS
3D structure
Package In Stock
Number of papers citing our products

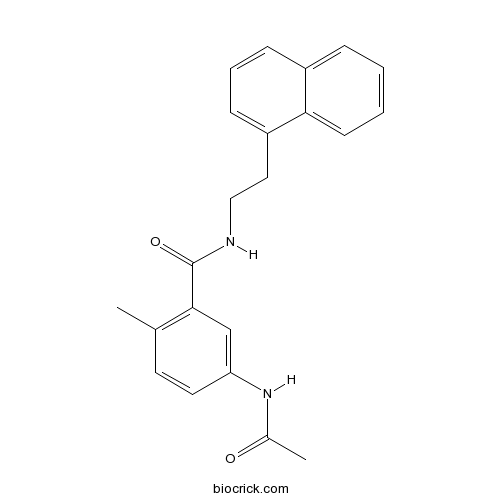
| Cas No. | 1093070-14-4 | SDF | Download SDF |
| PubChem ID | 44828572 | Appearance | Powder |
| Formula | C22H22N2O2 | M.Wt | 346.42 |
| Type of Compound | N/A | Storage | Desiccate at -20°C |
| Solubility | DMSO : ≥ 42 mg/mL (121.24 mM) *"≥" means soluble, but saturation unknown. | ||
| Chemical Name | 5-acetamido-2-methyl-N-(2-naphthalen-1-ylethyl)benzamide | ||
| SMILES | CC1=C(C=C(C=C1)NC(=O)C)C(=O)NCCC2=CC=CC3=CC=CC=C32 | ||
| Standard InChIKey | SSWAXHWHEZPTLC-UHFFFAOYSA-N | ||
| Standard InChI | InChI=1S/C22H22N2O2/c1-15-10-11-19(24-16(2)25)14-21(15)22(26)23-13-12-18-8-5-7-17-6-3-4-9-20(17)18/h3-11,14H,12-13H2,1-2H3,(H,23,26)(H,24,25) | ||
| General tips | For obtaining a higher solubility , please warm the tube at 37 ℃ and shake it in the ultrasonic bath for a while.Stock solution can be stored below -20℃ for several months. We recommend that you prepare and use the solution on the same day. However, if the test schedule requires, the stock solutions can be prepared in advance, and the stock solution must be sealed and stored below -20℃. In general, the stock solution can be kept for several months. Before use, we recommend that you leave the vial at room temperature for at least an hour before opening it. |
||
| About Packaging | 1. The packaging of the product may be reversed during transportation, cause the high purity compounds to adhere to the neck or cap of the vial.Take the vail out of its packaging and shake gently until the compounds fall to the bottom of the vial. 2. For liquid products, please centrifuge at 500xg to gather the liquid to the bottom of the vial. 3. Try to avoid loss or contamination during the experiment. |
||
| Shipping Condition | Packaging according to customer requirements(5mg, 10mg, 20mg and more). Ship via FedEx, DHL, UPS, EMS or other couriers with RT, or blue ice upon request. | ||

PLpro inhibitor Dilution Calculator

PLpro inhibitor Molarity Calculator
| 1 mg | 5 mg | 10 mg | 20 mg | 25 mg | |
| 1 mM | 2.8867 mL | 14.4333 mL | 28.8667 mL | 57.7334 mL | 72.1667 mL |
| 5 mM | 0.5773 mL | 2.8867 mL | 5.7733 mL | 11.5467 mL | 14.4333 mL |
| 10 mM | 0.2887 mL | 1.4433 mL | 2.8867 mL | 5.7733 mL | 7.2167 mL |
| 50 mM | 0.0577 mL | 0.2887 mL | 0.5773 mL | 1.1547 mL | 1.4433 mL |
| 100 mM | 0.0289 mL | 0.1443 mL | 0.2887 mL | 0.5773 mL | 0.7217 mL |
| * Note: If you are in the process of experiment, it's necessary to make the dilution ratios of the samples. The dilution data above is only for reference. Normally, it's can get a better solubility within lower of Concentrations. | |||||

Calcutta University

University of Minnesota

University of Maryland School of Medicine

University of Illinois at Chicago

The Ohio State University

University of Zurich

Harvard University

Colorado State University

Auburn University

Yale University

Worcester Polytechnic Institute

Washington State University

Stanford University

University of Leipzig

Universidade da Beira Interior

The Institute of Cancer Research

Heidelberg University

University of Amsterdam

University of Auckland

TsingHua University

The University of Michigan

Miami University

DRURY University

Jilin University

Fudan University

Wuhan University

Sun Yat-sen University

Universite de Paris

Deemed University

Auckland University

The University of Tokyo

Korea University
PLpro inhibitor is a potent inhibitor of papain-like protease (PLpro) with IC50 of 2.6 uM. IC50 Value: 2.6 uM [1] Target: PLpro; SARS-CoV in vitro: PLpro inhibitor is a potent inhibitor against the papain-like protease (PLpro) from the coronavirus that causes severe acute respiratory syndrome (SARS-CoV). PLpro inhibitor was found to have IC50 value of 2.6 ± 0.1 μM. PLpro inhibitor display significant antiviral activity with EC50 values of 13.1±0.7 uM, without toxicity up to the highest concentration tested. Notably, the increasing antiviral potency correlates with the in vitro inhibition of PLpro, suggesting that the compounds work directly on the enzyme in cells [1,2]. in vivo:
References:
[1]. Ratia, K., et al., A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc Natl Acad Sci U S A, 2008. 105(42): p. 16119-24.
[2]. http://www.google.com/patents/WO2010022355A1cl=en
- Angelol M
Catalog No.:BCN8271
CAS No.:1092952-64-1
- 7,2',4'-Trihydroxy-5-methoxy-3-phenylcoumarin
Catalog No.:BCN1626
CAS No.:1092952-62-9
- Ruxolitinib phosphate
Catalog No.:BCC1912
CAS No.:1092939-17-7
- Ruxolitinib sulfate
Catalog No.:BCC1913
CAS No.:1092939-16-6
- PP121
Catalog No.:BCC4980
CAS No.:1092788-83-4
- IT1t dihydrochloride
Catalog No.:BCC6234
CAS No.:1092776-63-0
- WAY 100635 hydrochloride
Catalog No.:BCC5061
CAS No.:146714-97-8
- (3S,4R)-Tofacitinib
Catalog No.:BCC4267
CAS No.:1092578-48-7
- (3S,4S)-Tofacitinib
Catalog No.:BCC4052
CAS No.:1092578-47-6
- (3R,4S)-Tofacitinib
Catalog No.:BCC4268
CAS No.:1092578-46-5
- Paxiphylline E
Catalog No.:BCN5886
CAS No.:1092555-03-7
- Paxiphylline D
Catalog No.:BCN5885
CAS No.:1092555-02-6
- B-Raf inhibitor 1
Catalog No.:BCC4182
CAS No.:1093100-40-3
- HNGF6A
Catalog No.:BCC8021
CAS No.:1093111-54-6
- 3-Hydroxy-4,15-dinor-1(5)-xanthen-12,8-olide
Catalog No.:BCN1625
CAS No.:1093207-99-8
- LKB1 (AAK1 dual inhibitor)
Catalog No.:BCC1705
CAS No.:1093222-27-5
- Vibralactone B
Catalog No.:BCN6748
CAS No.:1093230-95-5
- SRT2104 (GSK2245840)
Catalog No.:BCC1950
CAS No.:1093403-33-8
- VU 0155041
Catalog No.:BCC7615
CAS No.:1093757-42-6
- gamma-secretase modulator 2
Catalog No.:BCC1584
CAS No.:1093978-89-2
- CYM 5442 hydrochloride
Catalog No.:BCC7722
CAS No.:1094042-01-9
- SCH-1473759
Catalog No.:BCC1934
CAS No.:1094069-99-4
- Fmoc-His(Trt)-OH
Catalog No.:BCC3501
CAS No.:109425-51-6
- Fmoc-Orn(Boc)-OH
Catalog No.:BCC3533
CAS No.:109425-55-0
Inhibitor recognition specificity of MERS-CoV papain-like protease may differ from that of SARS-CoV.[Pubmed:25746232]
ACS Chem Biol. 2015 Jun 19;10(6):1456-65.
The Middle East Respiratory Syndrome coronavirus (MERS-CoV) papain-like protease (PLpro) blocking loop 2 (BL2) structure differs significantly from that of SARS-CoV PLpro, where it has been proven to play a crucial role in SARS-CoV PLpro inhibitor binding. Four SARS-CoV PLpro lead inhibitors were tested against MERS-CoV PLpro, none of which were effective against MERS-CoV PLpro. Structure and sequence alignments revealed that two residues, Y269 and Q270, responsible for inhibitor binding to SARS-CoV PLpro, were replaced by T274 and A275 in MERS-CoV PLpro, making critical binding interactions difficult to form for similar types of inhibitors. High-throughput screening (HTS) of 25000 compounds against both PLpro enzymes identified a small fragment-like noncovalent dual inhibitor. Mode of inhibition studies by enzyme kinetics and competition surface plasmon resonance (SPR) analyses suggested that this compound acts as a competitive inhibitor with an IC50 of 6 muM against MERS-CoV PLpro, indicating that it binds to the active site, whereas it acts as an allosteric inhibitor against SARS-CoV PLpro with an IC50 of 11 muM. These results raised the possibility that inhibitor recognition specificity of MERS-CoV PLpro may differ from that of SARS-CoV PLpro. In addition, inhibitory activity of this compound was selective for SARS-CoV and MERS-CoV PLpro enzymes over two human homologues, the ubiquitin C-terminal hydrolases 1 and 3 (hUCH-L1 and hUCH-L3).
A chimeric virus-mouse model system for evaluating the function and inhibition of papain-like proteases of emerging coronaviruses.[Pubmed:25100850]
J Virol. 2014 Oct;88(20):11825-33.
To combat emerging coronaviruses, developing safe and efficient platforms to evaluate viral protease activities and the efficacy of protease inhibitors is a high priority. Here, we exploit a biosafety level 2 (BSL-2) chimeric Sindbis virus system to evaluate protease activities and the efficacy of inhibitors directed against the papain-like protease (PLpro) of severe acute respiratory syndrome coronavirus (SARS-CoV), a biosafety level 3 (BSL-3) pathogen. We engineered Sindbis virus to coexpress PLpro and a substrate, murine interferon-stimulated gene 15 (ISG15), and found that PLpro mediates removal of ISG15 (deISGylation) from cellular proteins. Mutation of the catalytic cysteine residue of PLpro or addition of a PLpro inhibitor blocked deISGylation in virus-infected cells. Thus, deISGylation is a marker of PLpro activity. Infection of alpha/beta interferon receptor knockout (IFNAR(-/-)) mice with these chimeric viruses revealed that PLpro deISGylation activity removed ISG15-mediated protection during viral infection. Importantly, administration of a PLpro inhibitor protected these mice from lethal infection, demonstrating the efficacy of a coronavirus protease inhibitor in a mouse model. However, this PLpro inhibitor was not sufficient to protect the mice from lethal infection with SARS-CoV MA15, suggesting that further optimization of the delivery and stability of PLpro inhibitors is needed. We extended the chimeric-virus platform to evaluate the papain-like protease/deISGylating activity of Middle East respiratory syndrome coronavirus (MERS-CoV) to provide a small-animal model to evaluate PLpro inhibitors of this recently emerged pathogen. This platform has the potential to be universally adaptable to other viral and cellular enzymes that have deISGylating activities. Importance: Evaluating viral protease inhibitors in a small-animal model is a critical step in the path toward antiviral drug development. We modified a biosafety level 2 chimeric virus system to facilitate evaluation of inhibitors directed against highly pathogenic coronaviruses. We used this system to demonstrate the in vivo efficacy of an inhibitor of the papain-like protease of severe acute respiratory syndrome coronavirus. Furthermore, we demonstrate that the chimeric-virus system can be adapted to study the proteases of emerging human pathogens, such as Middle East respiratory syndrome coronavirus. This system provides an important tool to rapidly assess the efficacy of protease inhibitors targeting existing and emerging human pathogens, as well as other enzymes capable of removing ISG15 from cellular proteins.
Papain-like protease (PLpro) inhibitory effects of cinnamic amides from Tribulus terrestris fruits.[Pubmed:24882413]
Biol Pharm Bull. 2014;37(6):1021-8.
Tribulus terrestris fruits are well known for their usage in pharmaceutical preparations and food supplements. The methanol extract of T. terrestris fruits showed potent inhibition against the papain-like protease (PLpro), an essential proteolylic enzyme for protection to pathogenic virus and bacteria. Subsequent bioactivity-guided fractionation of this extract led to six cinnamic amides (1-6) and ferulic acid (7). Compound 6 emerged as new compound possessing the very rare carbinolamide motif. These compounds (1-7) were evaluated for severe acute respiratory syndrome coronavirus (SARS-CoV) PLpro inhibitory activity to identify their potencies and kinetic behavior. Compounds (1-6) displayed significant inhibitory activity with IC50 values in the range 15.8-70.1 microM. The new cinnamic amide 6 was found to be most potent inhibitor with an IC50 of 15.8 microM. In kinetic studies, all inhibitors exhibited mixed type inhibition. Furthermore, the most active PLpro inhibitors (1-6) were proven to be present in the native fruits in high quantities by HPLC chromatogram and liquid chromatography with diode array detection and electrospray ionization mass spectrometry (LC-DAD-ESI/MS).
Assessing activity and inhibition of Middle East respiratory syndrome coronavirus papain-like and 3C-like proteases using luciferase-based biosensors.[Pubmed:23986593]
J Virol. 2013 Nov;87(21):11955-62.
Middle East respiratory syndrome coronavirus (MERS-CoV) is associated with an outbreak of more than 90 cases of severe pneumonia with high mortality (greater than 50%). To date, there are no antiviral drugs or specific therapies to treat MERS-CoV. To rapidly identify potential inhibitors of MERS-CoV replication, we expressed the papain-like protease (PLpro) and the 3-chymotrypsin-like protease (3CLpro) from MERS-CoV and developed luciferase-based biosensors to monitor protease activity in cells. We show that the expressed MERS-CoV PLpro recognizes and processes the canonical CoV-PLpro cleavage site RLKGG in the biosensor. However, existing CoV PLpro inhibitors were unable to block MERS-CoV PLpro activity, likely due to the divergence of the amino acid sequence in the drug binding site. To investigate MERS-CoV 3CLpro activity, we expressed the protease in context with flanking nonstructural protein 4 (nsp4) and the amino-terminal portion of nsp6 and detected processing of the luciferase-based biosensors containing the canonical 3CLpro cleavage site VRLQS. Importantly, we found that a small-molecule inhibitor that blocks replication of severe acute respiratory syndrome (SARS) CoV and murine CoV also inhibits the activity of MERS-CoV 3CLpro. Overall, the protease expression and biosensor assays developed here allow for rapid evaluation of viral protease activity and the identification of protease inhibitors. These biosensor assays can now be used to screen for MERS-CoV-specific or broad-spectrum coronavirus PLpro and 3CLpro inhibitors.
Comparison of SARS and NL63 papain-like protease binding sites and binding site dynamics: inhibitor design implications.[Pubmed:22004941]
J Mol Biol. 2011 Nov 25;414(2):272-88.
The human severe acute respiratory syndrome coronavirus (SARS-CoV) and the NL63 coronaviruses are human respiratory pathogens for which no effective antiviral treatment exists. The papain-like cysteine proteases encoded by the coronavirus (SARS-CoV: PLpro; NL63: PLP1 and PLP2) represent potential targets for antiviral drug development. Three recent inhibitor-bound PLpro structures highlight the role of an extremely flexible six-residue loop in inhibitor binding. The high binding site plasticity is a major challenge in computational drug discovery/design efforts. From conventional molecular dynamics and accelerated molecular dynamics (aMD) simulations, we find that with conventional molecular dynamics simulation, PLpro translationally samples the open and closed conformation of BL2 loop on a picosecond-nanosecond timescale but does not reproduce the peptide bond inversion between loop residues Tyr269 and Gln270 that is observed on inhibitor GRL0617 binding. Only aMD simulation, starting from the closed loop conformation, reproduced the 180 degrees varphi-psi dihedral rotation back to the open loop state. The Tyr-Gln peptide bond inversion appears to involve a progressive conformational change of the full loop, starting at one side, and progressing to the other. We used the SARS-CoV apo X-ray structure to develop a model of the NL63-PLP2 catalytic site. Superimposition of the PLP2 model on the PLpro X-ray structure identifies binding site residues in PLP2 that contribute to the distinct substrate cleavage site specificities between the two proteases. The topological and electrostatic differences between the two protease binding sites also help explain the selectivity of non-covalent PLpro inhibitors.


